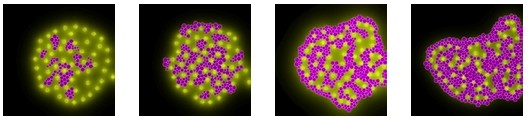
Cell aggregation (courtesy of Kurt Fleischer)
Time: TBD
Location: Remote Lecture Delivery, Regular Q&A Sessions
Optional Book:
The Origins of Life: From the Birth of Life to the Origin
of Language
by John Maynard Smith and Eors Szathmary
Students will carry out a number of programming projects during the course. These projects are designed to give students first-hand experience with a range of simulation methods that are used in biology. Grading for the class will primarily be based on student performance on these projects. Basic programming skills are recommended for students entering the course. Projects will be done using Processing, which is a Java-based programming environment. Prior programming experience in a Java or another C-like language is the best preparation for the course.
No previous background in biology is necessary.Homework 1 (Life Cellular Automata) - This first homework is due at the end of the second week of class.
Homework 2 (SIR Model of Infection)
Homework 3 (Reaction-Diffusion)
Homework 4 (Flocking)
Homework 5a (Mass/Spring Locomotion) 5b (Rigid Body Simulation). Pick just ONE of these for your last project!
Week 2
Coupled ordinary differential equations and numerical integration.
Agent-based models of interaction.
SIR epidemiological model of disease spread (susceptible, infectious,
recovered).
Week 3
Craig Reynolds'
flocking of virtual
creatures.
Week 4
Metabolic pathways chart1
and chart2.
Tim Hutton's self-reproducing simulated molecules.
Dill's 2D version of protein folding.
Optional:
Cyrus Levinthal
on protein folding complexity.
Classic DNA paper by
Watson and Crick.
Optional:
Stanley Miller's
bubbling flask to produce amino acids.
Seeing if
genetic code is optimal.
Week 5
Pattern formation by
reaction-diffusion.
Week 6
Simulation of
lipid micelles.
Week 9
Plant growth using
L-systems.
Creating branching patterns using
Laplacian growth.
Simulation of
leaf venation.
Week 10
Development papers:
Fleischer/Barr,
Eggenberger,
Furusawa/Kaneko.
Week 11
Evolving
virtual creatures
from Karl Sims.
Evolving
autonomous agents
from Frank Dellaert and Randall Beer.
Week 12
Evolution and manufacturing of
crawling robots.
Swimming creatures
from Tu and Terzopoulos.
Evolved flying creatures.
Week 13
Thomas Ray's Tierra
system of evolving programs.
Robert Axelrod and the
Iterated Prisoner's
Dilemma.
Week 14
Craig Reynolds on
Co-Evolution
for game
of tag.
Ant foraging
behavior.
Langton-style self-reproducing
loops from Hiroki Sayama.

Cell aggregation (courtesy of Kurt Fleischer)

Plant growth (courtesy of Przemyslaw Prusinkiewicz)
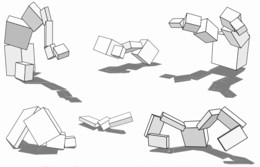
Walking simulation (courtesy of Karl Sims)
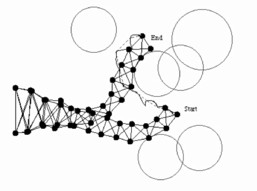
Tentacle motion (courtesy of Andrew Cantino)
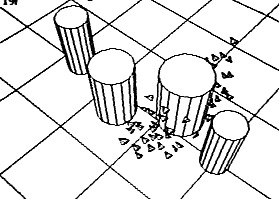
Flocking with collision avoidance (courtesy of Craig
Reynolds)
Go to Greg Turk's Home Page.