Project 2: Local Feature Matching

Trying to relate a dot in each image.
Accuracy of dots.
- Project Intro
- Code
- Results
- Conclusion
Project Intro
This project is a simplified version of the SIFT pipeline for local feature matching. This computes points that have a high likelihood to be corner points between two different images, due to the derivated Gaussian convolutions and the resultant high intesity points. It matches features that have been turned and a different orientation, ie, objects in different scenes and views. It detects interest points, finds features from the interest points, and matches them based on thresholds and distances.
Code
Point detection
This is described in Szeliski 4.1. A major part of this project is interest point detection, also known as key points detection. Shifted images is apt to provide large changes in intesity, helping to find key points. The first step is to filter the image with derivated guassian filters, in order to get the values needed to get the Harris value. This deals with editing the get_interest_points.m file. The small Gaussian filter is then convolved with a larger Gaussian, which would get the variables needed for the harris detection formula.
[Gx,Gy] = gradient(fspecial('gaussian',[3, 3], 0.5));
gx = imfilter(image,Gx);
gy = imfilter(image,Gy);
gxx = imfilter(gx,Gx);
gxy = imfilter(gy,Gx);
gyy = imfilter(gy,Gy);
Ixx = imfilter(gxx,fspecial('gaussian',[16 16],2));
Ixy = imfilter(gxy,fspecial('gaussian',[16 16],2));
Iyy = imfilter(gyy,fspecial('gaussian',[16 16],2));
The following satisfies the following variables in the harris formula:
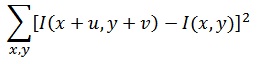
From there, we create the response matrix, which analyzes the 'cornerness' of each point given an intensity change. That code is demonstrated here, along with the application of the harris fomula:
responseMatrix = (Ixx.*Iyy)-Ixy.^2-0.06.*(Ixx+Iyy).^2;
responseMatrix(1 : 10, :) = 0;
responseMatrix(end - 10 : end, :) = 0;
responseMatrix(:, 1 : 10) = 0;
responseMatrix(:, end - 10 : end) = 0;
The following code shows the response sorting of the response matrix.
% sorting the responses to get the best keypoints
index = imregionalmax(responseMatrix);
sortedResp = sort(responseMatrix(index),'descend');
keyPoints = 1000;
topResp = sortedResp(1:keyPoints);
x=[];y=[];
for aPoint = 1:keyPoints
[row,col]=find(responseMatrix==topResp(aPoint));
x=[x; col];
y=[y; row];
end
Feature description
The SIFT pipeline next builds local feature descriptors. First, we divide up a 16x16 grid around each point. We then comput the gradient histogram with a gaussian filter. We sum up the magnitudes according to orientation, and then do some normalization to make it easy to process by Matlab. This is done in Szeliski 4.1.2.
% Creating the guassian filters
gaussian = fspecial('gaussian', [feature_width, feature_width], 1);
convolvedGauss = fspecial('gaussian', [feature_width, feature_width], feature_width / 2);
[GaussDx, GaussDy] = gradient(gaussian);
% Apply the guassian filter
Ix = double(imfilter(image, GaussDx));
Iy = double(imfilter(image, GaussDy));
% magnitude and direction calculations
magnitude = sqrt(Ix .^ 2 + Iy .^ 2);
direction = mod(round((atan2(Iy, Ix) + 2 * pi) / (pi / 4)), 8);
% Going over xy pairs
for aPoint = 1 : length(x)
% Center of the grid
xCenter = x(aPoint);
yCenter = y(aPoint);
% Offset of feature witdth on the centers
gridSize = feature_width / 4;
xOffset = xCenter - gridSize * 2;
yOffset = yCenter - gridSize * 2;
% Smooth the magitude matrix
bigMagnitudeGrid = magnitude(yOffset: yOffset + feature_width - 1, xOffset: xOffset + feature_width - 1);
bigDirectionGrid = direction(yOffset: yOffset + feature_width - 1, xOffset: xOffset + feature_width - 1);
bigMagnitudeGrid = imfilter(bigMagnitudeGrid, convolvedGauss);
% Iterating over the image
for xi = 0 : 3
for yi = 0 : 3
directionGrid = bigDirectionGrid((gridSize * xi + 1) : (gridSize * xi + gridSize), ...
(gridSize * yi + 1) : (gridSize * yi + gridSize));
magnitudeGrid = bigMagnitudeGrid((gridSize * xi + 1) : (gridSize * xi + gridSize), ...
(gridSize * yi + 1) : (gridSize * yi + gridSize));
for d = 0 : 7
mask = directionGrid == d;
features(aPoint, (xi * 32 + yi * 8) + d + 1) = sum(sum(magnitudeGrid(mask)));
end
end
end
% Normalization of the features
magnitudeSum = sum(features(aPoint, :));
features(aPoint, :) = features(aPoint, :) / magnitudeSum;
end
Feature Matching
For match_features.m, the feature matching is done using the euclidean ratio test and Szeliski 4.1.3. First, we get the distance matrix between the set of key points of the two images. Next, sort the distance matrix by the rows. Then, the rows with ratios of the nearest neighbors and second nearest neighbors beyond a threshold are used. The distance is how similarity of the features are, and the ratio of the smallest distance and the second small one is the reliability.
threshold = 0.6;
k = 1;
confidences = [];
matches = [];
for i = 1 : size(features1, 1)
distances = features2;
for j = 1 : size(distances, 1)
distances(j, :) = distances(j, :) - features1(i, :);
distances(j, :) = distances(j, :) .^ 2;
end
[distList, index] = sort(sum(distances, 2));
confidence = distList(2) / distList(1);
if threshold > 1 / confidence
matches(k, 1) = i;
matches(k, 2) = index(1);
confidences(k) = confidence;
k = k + 1;
end
end
Results


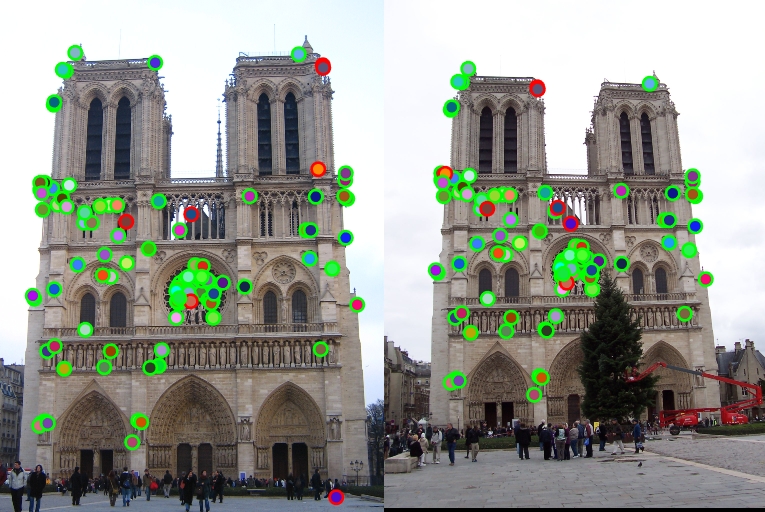
Notre Dame matched 117 out of 124 dots correctly, with 7 incorrectly for 94% accuracy. |



Mount Rushmore matched 31 out of 35 dots correctly, with 4 incorrectly for 89% accuracy. |



Episcopal Gaudi matched 0 out of 4 dots correctly, with 4 incorrectly for 0% accuracy. |
Conclusion
Some of my results are passable, such as Notre Dame and Mount Rushmore. The Episcopal Gaudi received terrible results, missing all of the points. Messing with threshholds has adjusted the results - for example, increasing the threshold in match_features.m reduced accuracy by 10%. The results are not perfect, but do fairly okay within a certain space. Changing threshold values adjusts the results, so I probably could have gotten a better score on the Episocpal Gaudi.