Project 2: Local Feature Matching
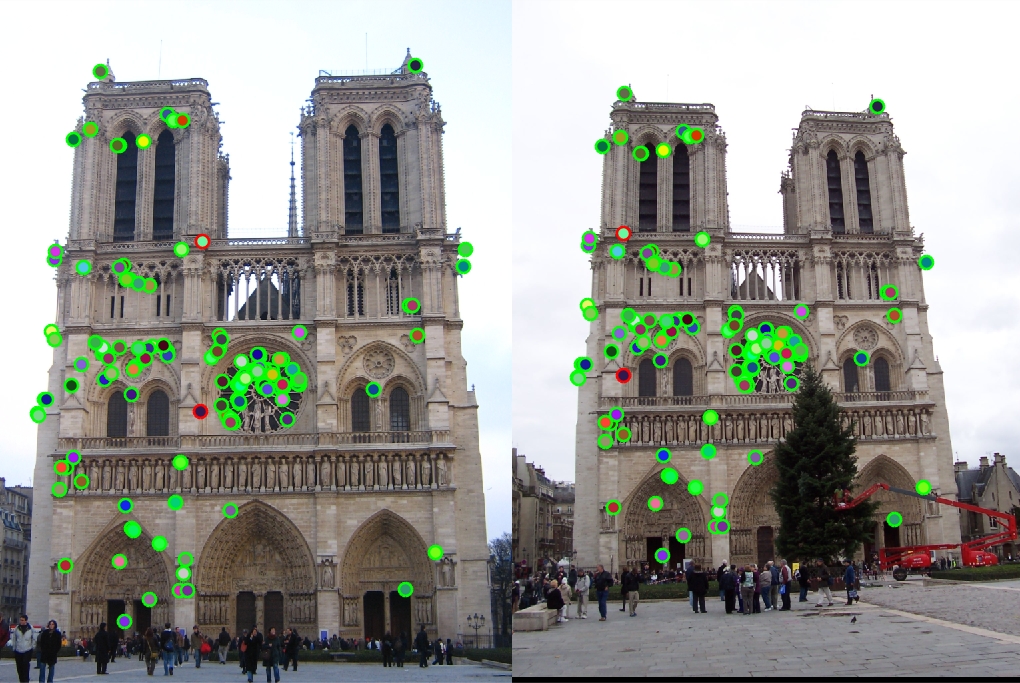
Example of a local feature matches.
The purpose of this project is to create a local feature matching algorithm implementing SIFT pipeline. The matching pipeline is built to work for instance-level matching multiple views of the same physical scene. For this project, three major steps of a local feature matching algorithm are implemented:
- Interest point detection
- Local feature description
- Feature matching
An example of local feature matches from a baseline implementation of this project is shown on the right.
Code and Algorithms
Interes Point Detection (get_interest_points.m)
The code shown below implements the Harris corner detector. The baseline Harris corner detector algorithm is:
- Compute the horizontal and vertical derivatives of the image Ix and Iy by convolving the original image with derivatives of Gaussians
- Compute the three images corresponding to the outer products of these gradients. (The matrix A is symmetric, so only three entries are needed.)
- Convolve each of these images with a larger Gaussian
- Compute a scalar interest
- Find local maxima above a certain threshold and report them as detected feature point locations.
function [x, y, confidence, scale, orientation] = get_interest_points(image, feature_width)
[imagerow,imagecolumn]=size(image);
diff_Gaussian=fspecial('Gaussian',2)-fspecial('Gaussian',1);% Pre-defined Gaussian Matrix for differente dimensions
four_dim_Gaussian=fspecial('gaussian',4);
dx=[-1,0,1];
dy=[-1;0;1];
Fx=conv2(image,dx,'same');% Image derivatives: Calculate the derivatives fx
Fy=conv2(image,dy,'same');% Image derivatives:Calculate the derivatives fy
F_x=imfilter(Fx,diff_Gaussian);
F_y=imfilter(Fy,diff_Gaussian);
Fx_square=imfilter(F_x.^2,four_dim_Gaussian);%Smoothed squared image derivatives
Fy_square=imfilter(F_y.^2,four_dim_Gaussian);
Fxy=imfilter(F_x.*F_y,four_dim_Gaussian);
harris=Fx_square.*Fy_square-Fxy.^2-0.04*((Fx_square+Fy_square).^2);%Calculate the harris value
maxima=colfilt(harris,[3,3],'sliding',@max);% find local maxima
x=[];
y=[];
for a=1:imagerow
for b=1:imagecolumn
if (harris(a,b)==maxima(a,b)&& (harris(a,b)>0.001))% Free Parameter: interest response threshold
x=vertcat(x,b);
y=vertcat(y,a);
end
end
end
end
Local Feature Description (get_features.m)
In this part, a SIFT-like local feature is implemented. More specifically, it returns a set of feature descriptors for a set of interest points given by the previous sub-function. In this project, the parameter “feature width” is set to be a constant 16. The code below firstly calculates the gradient magnitude and direction of the given image. Then, the image is filtered by a Gaussian filter. A 16 by 16 window around the detected keypoint and a gradient orientation histogram are created by adding the weighted gradient value to eight orientation histogram bins. The resulting 128 feature vectors are normalized to unit length. At last, the feature vectors are once again normalized to unit length after doing the threshold calculation.
function [features] = get_features(image, x, y, feature_width)
[imagerow,imagecolumn]=size(image);
[Gmag,Gdir] = imgradient(imfilter(image,fspecial('gaussian')));%calculate Gradient magnitude and direction
Gmag=imfilter(Gmag,fspecial('gaussian'));
Gdir=imfilter(Gdir,fspecial('gaussian'));
features=[];
for n=1:size(x)%iterate every interest point
histograms=zeros(4,4,8);% 4*4*8 histograms
cell_first_row=y(n)-8;
cell_last_row=y(n)+feature_width-1-8;
cell_first_column=x(n)-8;
cell_last_column=x(n)+feature_width-1-8;
if ( cell_first_row>0 && cell_last_row<=imagerow && cell_first_column>0 && cell_last_column <=imagecolumn )
direction=mat2cell(Gdir(cell_first_row:cell_last_row,cell_first_column:cell_last_column),[4 4 4 4],[4 4 4 4]);
magnitude=mat2cell(Gmag(cell_first_row:cell_last_row,cell_first_column:cell_last_column),[4 4 4 4],[4 4 4 4]);
for a=1:4
for b=1:4
for new_row=1:4
for new_column=1:4
if( direction{a,b}(new_row,new_column)>=0&&direction{a,b}(new_row,new_column)<45 )
histograms(a,b,1)=histograms(a,b,1)+magnitude{a,b}(new_row,new_column);
end
if(direction{a,b}(new_row,new_column)>=45&&direction{a,b}(new_row,new_column)<90)
histograms(a,b,2)=histograms(a,b,2)+magnitude{a,b}(new_row,new_column);
end
if(direction{a,b}(new_row,new_column)>=90&&direction{a,b}(new_row,new_column)<135)
histograms(a,b,3)=histograms(a,b,3)+magnitude{a,b}(new_row,new_column);
end
if(direction{a,b}(new_row,new_column)>=135&&direction{a,b}(new_row,new_column)<180)
histograms(a,b,4)=histograms(a,b,4)+magnitude{a,b}(new_row,new_column);
end
if(direction{a,b}(new_row,new_column)>=-45&&direction{a,b}(new_row,new_column)<0)
histograms(a,b,5)=histograms(a,b,5)+magnitude{a,b}(new_row,new_column);
end
if(direction{a,b}(new_row,new_column)>=-90&&direction{a,b}(new_row,new_column)<-45)
histograms(a,b,6)=histograms(a,b,6)+magnitude{a,b}(new_row,new_column);
end
if(direction{a,b}(new_row,new_column)>=-135&&direction{a,b}(new_row,new_column)<-90)
histograms(a,b,7)=histograms(a,b,7)+magnitude{a,b}(new_row,new_column);
end
if(direction{a,b}(new_row,new_column)>=-180&&direction{a,b}(new_row,new_column)<-135)
histograms(a,b,8)=histograms(a,b,8)+magnitude{a,b}(new_row,new_column);
end
end
end
end
end
end
fvector=reshape(histograms,[1,128])./norm(reshape(histograms,[1,128]));% build 128 D vectors and each feature should be normalized to unit length
fvector(fvector>0.8)=0;%Free Parameters: feature vector threshold
f_final=fvector./norm(fvector);% normalize again
features=vertcat(features,f_final.^0.6);%raise each element of the final feature vector to some power that is less than one
end
end
Feature Matching (match_features.m)
In feature matching part, the nearest neighbor distance ratio test method of matching local features is implemented. To begin with, the Euclidean (vector magnitude) distances in feature space is directly used for ranking potential matches. Then NNDR is calculated to get the matches.
function [matches, confidences] = match_features(features1, features2)
features1_length=size(features1,1);
features2_length=size(features2,1);
matches=[];
confidences=[];
for a=1:features1_length
for b=1:features2_length
distance(a,b)=sqrt(sum((features1(a,:)-features2(b,:)).^2));%euclidean distance
end
end
for a=1:features1_length
sort_distance=sort(distance(a,:));
NNDR=sort_distance(1)/sort_distance(2);%nearest and second nearest neighbor distances
match_distance=find(distance(a,:)==sort_distance(1));
if(NNDR<0.8) % Free parameter: nearest neighbor distance ratio
matches=vertcat(matches,[a,match_distance(1)]);
confidences=vertcat(confidences,1/NNDR);
end
end
% Sort the matches so that the most confident onces are at the top of the
% list. You should probably not delete this, so that the evaluation
% functions can be run on the top matches easily.
[confidences, ind] = sort(confidences, 'descend');
matches = matches(ind,:);
Results and Discussion
Notre Dame Results
 |
 |
 |
Feature Vector Threshold = 0.5, NNDR = 0.9, 934 good matches, 323 bad matches, 74% accuracy
 |
 |
 |
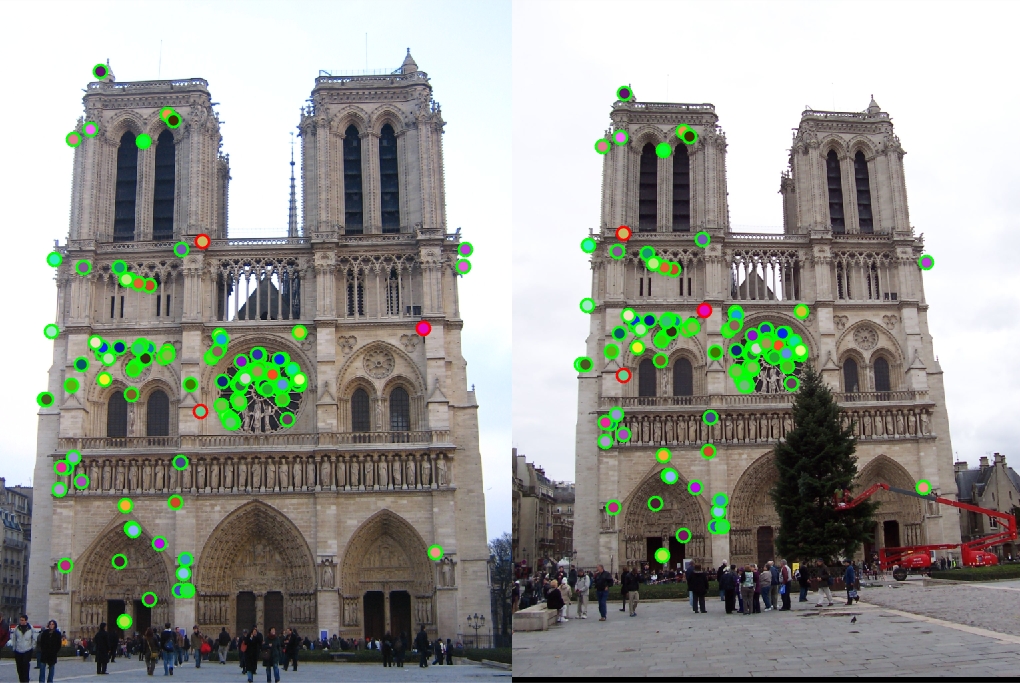
Feature Vector Threshold = 0.5, NNDR = 0.7, 122 good matches, 3 bad matches, 98% accuracy
 |
 |
 |
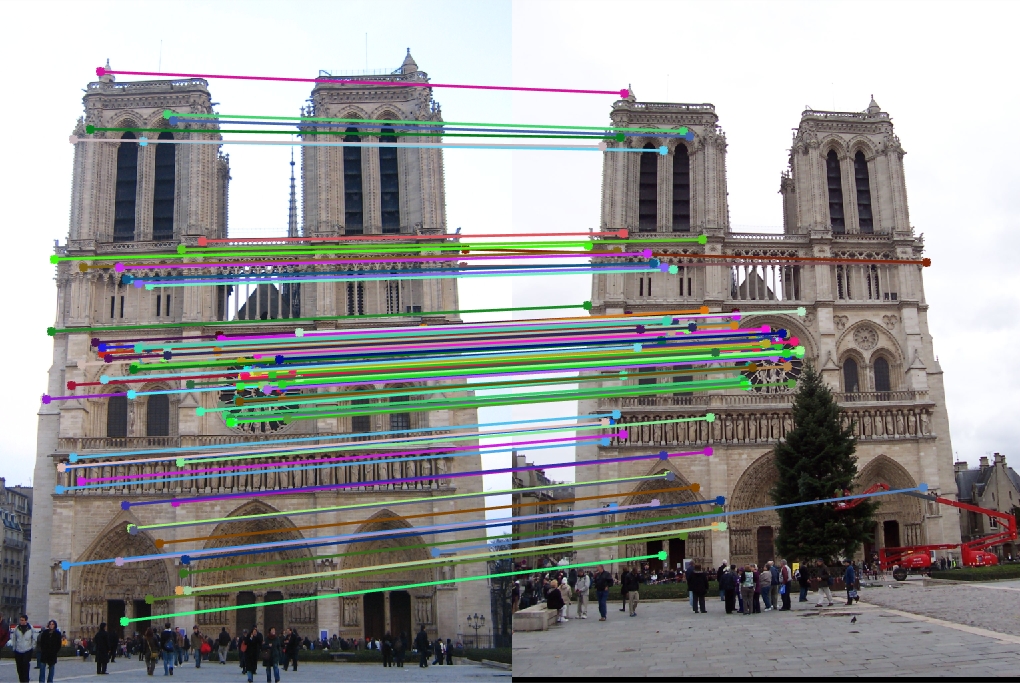
Feature Vector Threshold = 0.8, NNDR = 0.7, 137 good matches, 2 bad matches, 99% accuracy
Mount Rushmore Results
 |
 |
 |
Feature Vector Threshold = 0.5, NNDR = 0.9, 765 good matches, 97 bad matches, 89% accuracy
 |
 |
 |
Feature Vector Threshold = 0.5, NNDR = 0.7, 85 good matches, 1 bad matches, 99% accuracy
 |
 |
 |
Feature Vector Threshold = 0.8, NNDR = 0.7, 108 good matches, 1 bad matches, 99% accuracy
Episcopal Gaudi Results
 |
 |
 |
Feature Vector Threshold = 0.5, NNDR = 0.9, 5 good matches, 50 bad matches, 9% accuracy
 |
 |
 |
Feature Vector Threshold = 0.8, NNDR = 0.8, 3 good matches, 2 bad matches, 60% accuracy
 |
 |
 |
Feature Vector Threshold = 0.8, NNDR = 0.9, 7 good matches, 57 bad matches, 11% accuracy
Some Extra Works
 |
 |
GOOD: Image Pair: House. Feature Vector Threshold = 0.8, NNDR = 0.8
 |
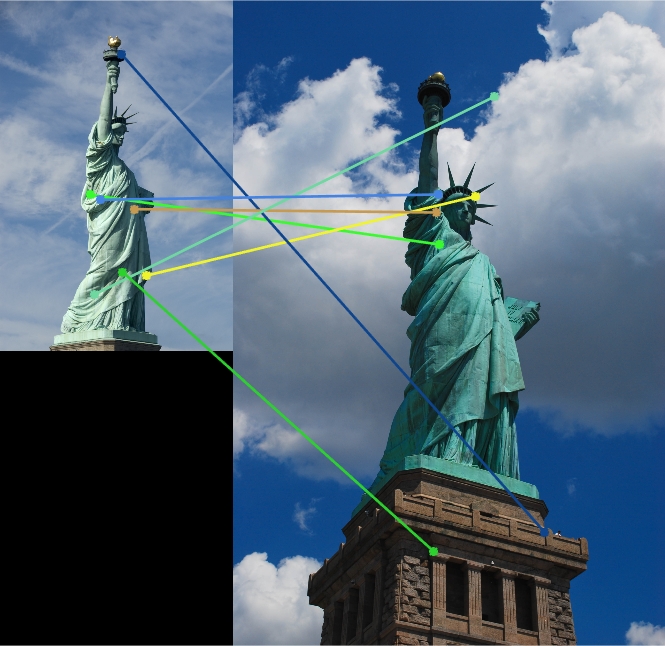 |
BAD: Image Pair: Statue of Liberty.Feature Vector Threshold = 0.8, NNDR = 0.8
In this part, several pairs of image in extradata file are used to test the above three functions. Although there are no eval (ground truth) file to illustrate the accuracy, it can be found that for House Image, the accuracy is quite high. The reason could be that the original image pair is very similar so it will not be hard to match certain interest points. However, for the below image pair Statue of Liberty, the accuracy, by observing, is relatively low. The reason can be the threshold value for finding interest points and the threshold value for matching features (NNDR) are too high for this image pair.
Discussion
In this project, for the Notre Dame image pair, first of all, the accuracy is relatively low. Then, according to the instruction in get_features.m, each element of the final feature vector is raised to the power of 0.6. Then, the feature vector threshold is set to be 0.5 and NNDR is set to be 0.9. The result, as shown above, generates a large amount of interest points and the accuracy is still not satisfactory. Next, the NNDR is decreased to 0.7 to reduce the number of interest point pairs. For this time, the result gets 122 good matches and 3 bad matches, and the accuracy increases to 98%. For the third time, the NNDR remains 0.7 but the feature vector threshold is increased to 0.8 in order to increase the amount of interest points. The accuracy gets higher to 99%. For the image pair of Mount Rushmore, the similar results can be achieved. For Episcopal Gaudi, when the Feature Vector Threshold is set to be 0.8 and NNDR is set to be 0.8, the result gets 3 good matches and 2 bad matches. The result is not applicable since there is not enough interest points. When the feature vector threshold is set to be 0.8 and NNDR is set to be 0.9, the accuracy increases to 11%, with 7 good matches and 57 bad matches.