Project 2: Local Feature Matching
Content
1. Overview
A local feature matching algorithm on instance-level is implemented in this assignment to match same physical scenes from multiple
views. Using simplified versions of the famous SIFT pipeline, the algorithm could first detect interest points in get_interest_points.m
. Then the local feature is described in each interest point in get_features.m. Finally, feature matching is realized
in match_features.m. Simple implementations are experimented first and then more sophisticated algorithms are adapted to increase
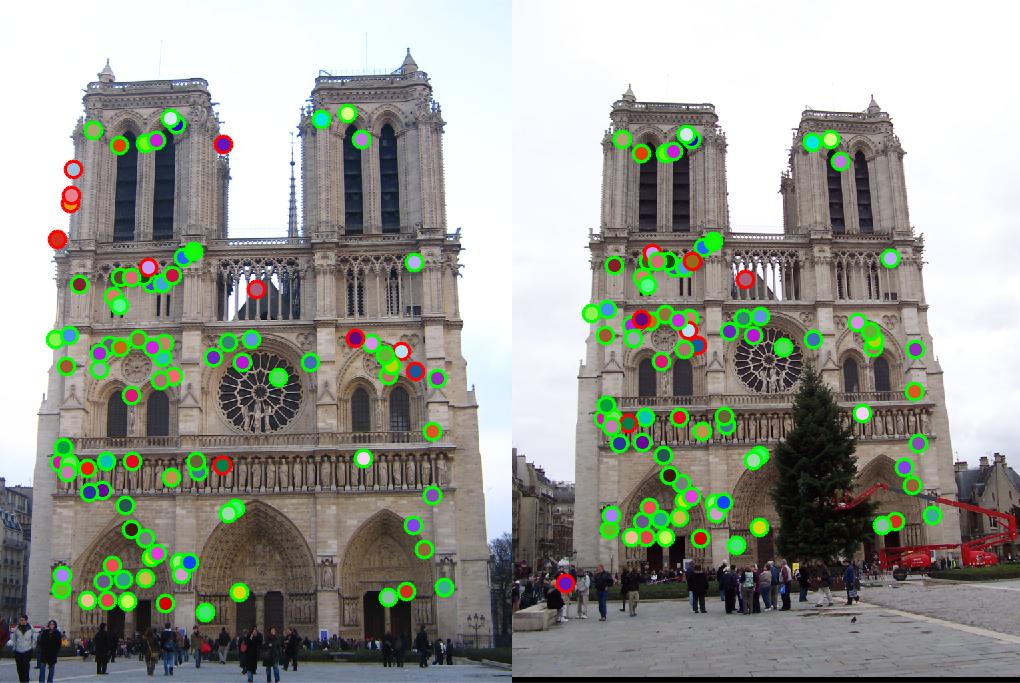
the successful match rate. Fig. 1 shows the Notre Dame image pair on which we will match features.

|

|
2. Implementation
- Interest point detection
Harris corner detector is implemented in get_interest_points.m. The second moment matrix is used to approximate local
auto-correlation which represents distinctiveness. To calculate the second moment matrix, Sobel-like filter is used to calculate the gradients
in both x- and y- directiones. A gaussian filter is applied on each element of the matrix.
[rows,cols] = size(image);
Xfilter = [-1,0,1;-1,0,1;-1,0,1];
Yfilter = Xfilter';
Ix = imfilter(image,Xfilter);
Iy = imfilter(image,Yfilter);
gauss = fspecial('gaussian');
Ix2 = imfilter(Ix.^2, gauss);
Iy2 = imfilter(Iy.^2, gauss);
Ixy = imfilter(Ix.*Iy,gauss);
alpha = 0.06. Then Matlab colfilt function is
applied to assign local maximum score to each pixel. The local window size is determined by feature_width, which is an
input value for this function.
alpha = 0.06;
harris = (Ix2.*Iy2 - Ixy.^2) - alpha*((Ix2+Iy2).^2);
maxi = colfilt(harris,[feature_width,feature_width],'sliding',@max);
threshold = 0.0003;
extracted = (harris==maxi) & (harris > threshold);
boundaryX = 5*feature_width;
boundaryY = 5*feature_width;
[y, x] = find(extracted(boundaryY:rows-boundaryY,boundaryX:cols-boundaryX));
y = boundaryY + y - 1;
x = boundaryX + x - 1;
- Local feature description
A SIFT-like local feature description is realized in get_features.m. Given an interest point, locally we find a 16x16 matrix and divide it
into 4x4 section equally. For each section, the gradients are distributed over a histogram on gradient directions. The histogram is
chosen to be of size 8. Then at each interest point, we have a 4x4x8 = 128 dimensional vector.
As the interest point could be at the boundaries in general cases (not in get_interest_points.m), the input image is
padded first. imgradient function is used to calculate the image gradients Gmag and directions Gdir
on pixel level. Local gradient matrix and gradient direction matrix are stored in local and local_dir respectively.
Then histograms at all the sections are calculated.
half = feature_width/2;
quad = feature_width/4;
image = padarray(image,[half,half]);
[Gmag,Gdir] = imgradient(image);
x = x + half;
y = y + half;
dim_x = size(Gmag,2);
dim_y = size(Gmag,1);
for i = 1:length(y)
x(i) = round(x(i));
y(i) = round(y(i));
local = Gmag(y(i)-half:y(i)+half-1,x(i)-half:x(i)+half-1);
local_dir = Gdir(y(i)-half:y(i)+half-1,x(i)-half:x(i)+half-1);
current_feature = [];
for j = 0:3
for k = 0:3
temp_grad = local(1+quad*j:quad*(j+1),1+quad*k:quad*(k+1));
temp_dir = local_dir(1+quad*j:quad*(j+1),1+quad*k:quad*(k+1)) + 180;
temp_grad = reshape(temp_grad,[16,1]);
temp_dir = reshape(temp_dir,[16,1]);
temp_dir = floor(temp_dir/45)+1;
hist_count = zeros(8,1);
for count = 1:16
hist_count(temp_dir(count),1) = hist_count(temp_dir(count),1) + temp_grad(count);
end
hist_count = hist_count/sum(hist_count);
current_feature = [current_feature;hist_count];
end
end
coord = [x(i)/dim_x;y(i)/dim_y]*coord_factor;
current_feature = [current_feature;coord];
current_feature = current_feature/norm(current_feature,2);
features(i,:) = current_feature';
end
- Feature Matching
Given two sets of features extracted from images of two different views, we would want to match features across the images.
Each feature is a high dimensional vector (128 dimensional for abovementioned algorithm). Euclidean distance is the only used metric.
For each feature in image 1, all the features in image 2 would be iterated to calculate the Euclidean distances to the feature in image
1 and save the distances in a vector called feature_dist. The feature in image 2 which has the smallest distance nearest
to the feature in image 1 is the match. And the second smallest distance second_nearest is used to calculate the confidence.
If second_nearest is much bigger than nearest, then the matched feature is more likely to be a right match.
Thus confidence is calculated by 1-nearest/second_nearest. Sorting the matches based on the confidence scores
we can select matches with higher confidence scores. With generating more than 100 features in each image, top 100 matches will be
selected to be the output for match_features.m.
num_features = min(size(features1, 1), size(features2,1));
matches = zeros(num_features, 2);
AllPossMatch = [];
for i = 1:size(features1,1)
feature_dist = [];
for j = 1:size(features2,1)
feature_dist =[feature_dist; norm(features1(i,:) -features2(j,:))];
end
[nearest,pos] = min(feature_dist);
feature_dist(pos) = [];
second_nearest = min(feature_dist);
confidence = 1 - nearest/second_nearest;
AllPossMatch = [AllPossMatch;[i,pos,confidence]];
end
confidences = AllPossMatch(:,3);
[confidences, ind] = sort(confidences, 'descend');
sorted_matches = AllPossMatch(ind,1:2);
matches = sorted_matches(1:num_features,:);
matches = matches(1:100,:);

|

|
|
3. Results and Discussion
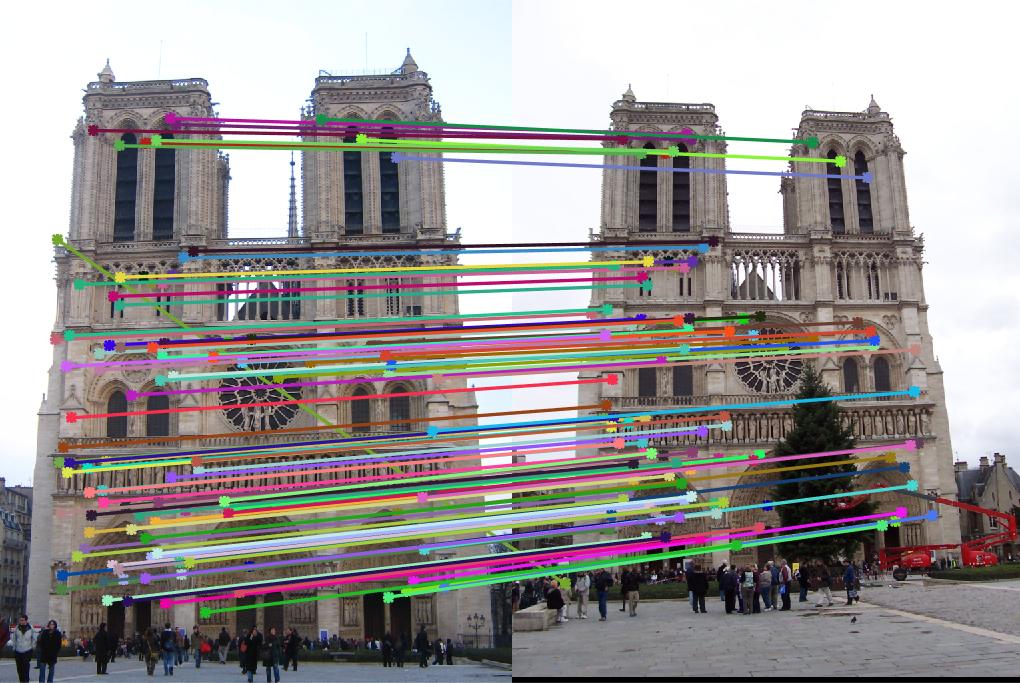
Using the abovementioned algorithms, we first match the features in the Notre Dame image pair and reach to 88% accuracy using
threshold = 0.0003 in get_interest_points.m. This results is shown in Fig. 2 taking as baseline.
Lowering the threshold = 0.000001 will also increase the match accuracy up to 2% (90%). Note that, bad matches actually happens in the case that
multiple features in image 1 are matched to the same feature in image 2, which is not ideal. Feature should be matched individually,
meaning each feature should have only one match. To resolve this issue, each multiple matched feature should be detected. And the according
best match (with highest confidence) is saved. The other misleading match indexes are saved to abandonList and then got
deleted in the AllPossMatch matrix.
abandonList = [];
[match2,index] = sort(AllPossMatch(:,2),'descend');
maxConf = 0;maxIndex = 1;
for i = 2:size(match2)
if match2(i) == match2(i-1)
maxConf = max([maxConf,AllPossMatch(index(i),3), AllPossMatch(index(i-1),3)]);
if maxConf > max([AllPossMatch(index(i),3), AllPossMatch(index(i-1),3)])
abandonList = [abandonList,index(i)];
elseif maxConf == AllPossMatch(index(i),3)
abandonList = [abandonList,index(maxIndex)];
maxIndex = i;
else
abandonList = [abandonList,index(i)];
end
else
maxConf = 0;
maxIndex =i;
end
end
AllPossMatch(abandonList,:) = [];
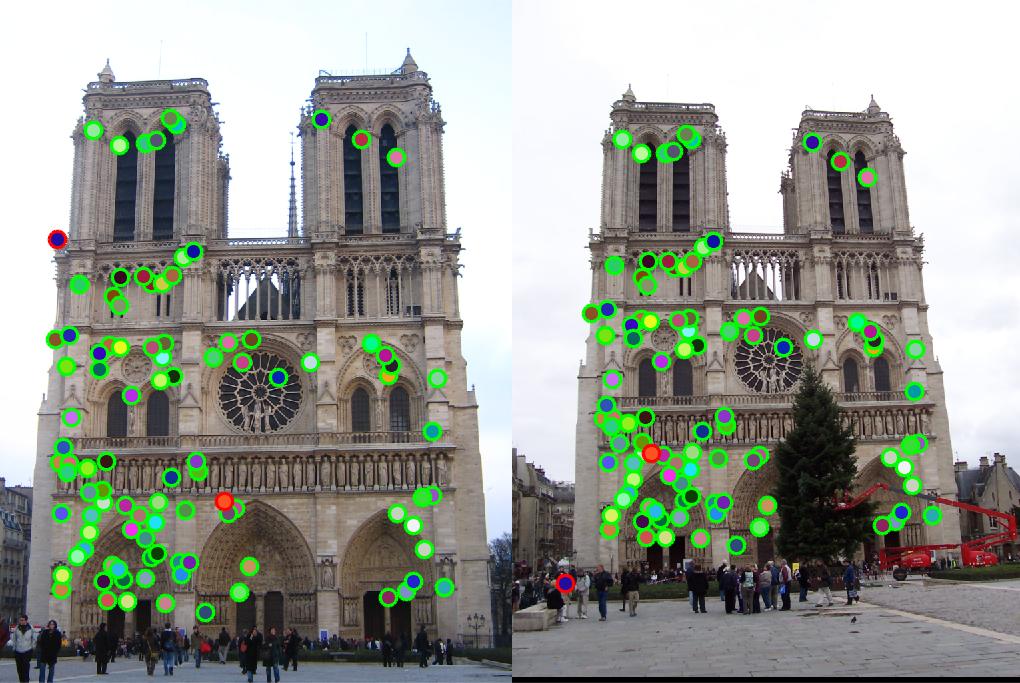
match_features.m, the accuracy reaches to 98%. Fig. 3 shows the matching results.

|
|
|
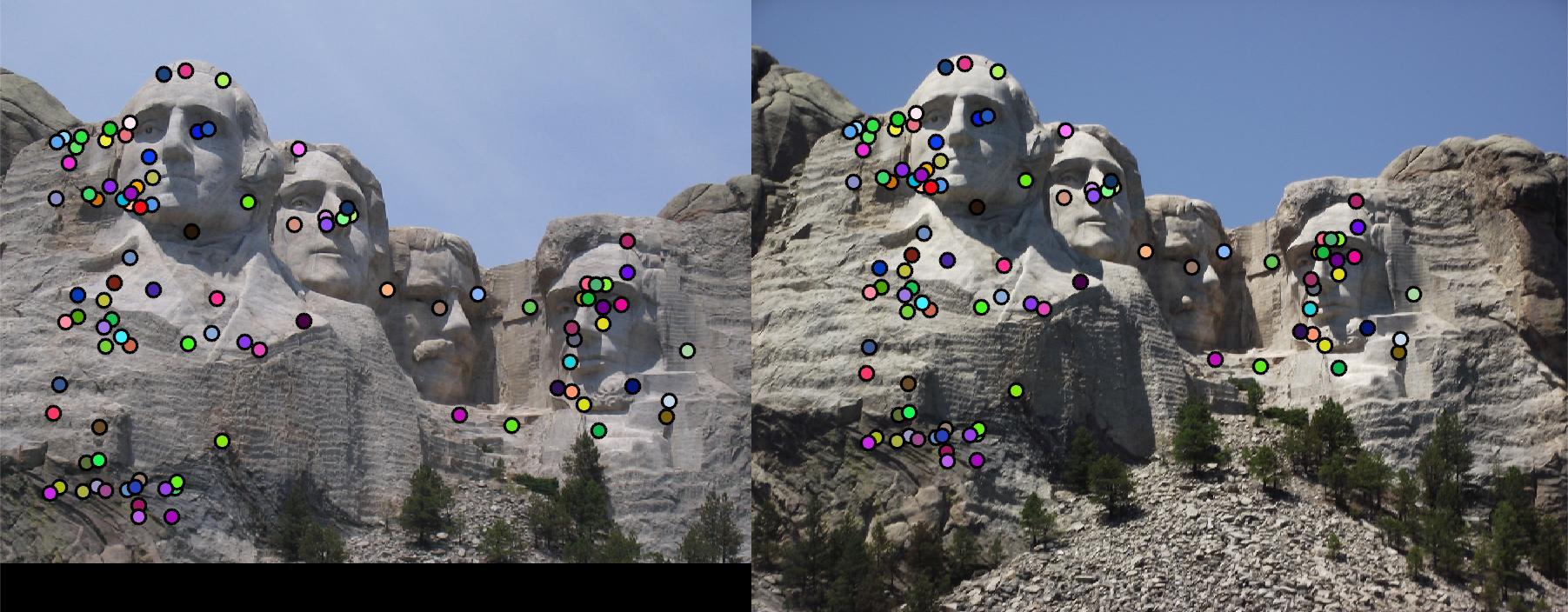
Let threshold = 0.0003, and use the same code to run feature matching on Mount Rushmore pair. 92% accuracy is achieved.
Fig. 4 shows the match result. However, due to strong similarity of the corners in Episcopal Gaudi pair, accuracy is quite low,
results of which show in Fig. 5.

|

|

|

|

|

|
4. Extra credits attempted
As the baseline implementation results high accuracy for Notre Dame pair already, the Mount Rushmore pair will be used for examining the extra credit.
- Keypoint Orientation Estimation: 5pts
When detecting interest point in Harris corner detector, we can also estimate local orientation for each
interst point. Saving the local orientation in vector orientation, we can further generate local
feature rotation invariant. ls is the local orientation calculator size. For the Mount Rushmore pair,
20 is the relative good size, increasing the accuracy from 92% to 97%. The full implementation is in get_interest_points_extra.m
.
orientation = zeros(length(x),1);
ls = 20;
[Gmag,Gdir] = imgradient(image);
for i = 1:length(x)
local_ori_matrix = Gdir(y(i)-ls:y(i)+ls-1,x(i)-ls:x(i)+ls-1);
orientation(i) = sum(sum(local_ori_matrix))/(2*ls)/(2*ls);
end
- Local Feature Descriptor with rotation invariance: 5pts
As get_interest_points_extra.m can estimate orientation, the local feature descriptor should also have
rotational invariance. Simply add the orientation at each interest point to the local gradient direction matrix, and then
limit the range of local direction to [0,2pi]. Full implementation is in get_features_extra.m. Fig. 6 shows
the Mount Rushmore pair using this local feature descriptor with rotation invariance. Accuracy is increased to 97%.
local_dir = local_dir + orientation(i);
local_dir = local_dir + 360*(local_dir<0);
local_dir = local_dir - 360*(local_dir>360);
|

|

|
- Adaptive Non-maximum Suppression (ANMS): 5pts
In case that interest points can be denser in regions of higher contrast, ANMS detect features that are not
only local maximum, but also have cornerness score significantly higher (percentage determined by ANMS_diff
)than all its neighbors within a radius. Full implementation is in get_interest_point_ANMS.m.
Using this ANMS interest point detector and regular baseline feature descriptor, the Mount Rushmore pair get 97%
accuracy, which is 5% higher than the regular interest point detector.
copyX = x;
copyY = y;
x = [];y =[];
ANMS_diff = 0.1;
radius = 0.25*feature_width;
for i = 1:length(copyY)
local = gray_harris(copyY(i)-radius:copyY(i)+radius,copyX(i)-radius:copyX(i)+radius);
local = reshape(local,[(radius*2+1)^2,1]);
current = gray_harris(copyY(i),copyX(i));
local(local == current) =[];
if current*(1-ANMS_diff) > max(local)
x = [x;copyX(i)];
y = [y;copyY(i)];
end
end
- Gradient location-orientation histogram (GLOH) spatial layouts: 5pts
GLOH is a variant on SIFT. Instead of using square local sections, GLOH uses log-polar bins to computer orientation
histogram. Using radius of 6, 11, 15 to divide regions, get_features_GLOH.m generates 17 sections, each of
which has a histogram of 8 bins. The feature size is 17*8. With baseline interst point detector and the GLOH feature
extractor, 99% accuracy has been easily achieved on Mount Rushmore pair. The match results are shown in Fig. 7.

|

|

|