Project 3 / Camera Calibration and Fundamental Matrix Estimation with RANSAC
Camera Projection Matrix
I computed the camera projection matrix that goes from the world 3D coordinates to 2D image coordinates using homogeneous coordinates. Using this matrix (M), I then calculated the camera center for both the sets of points given (normalised and unnormalised.)
My
- Step 1: Setup a homogenous set of equations using the corresponding 2D and 3D points (Ax = 0)
- Step 2: Find the SVD of matrix A
- Step 3: Compute M using the following commands:
[U, S, V]= svd(A);
M = V(:,end);
M = reshape(M,[],3)';
- Step 4: Now compute the camera center = -1*inv(Q)/m4 where Q = left 3x3 matrix of M and m4 = 4th column of M
Results:
|
|
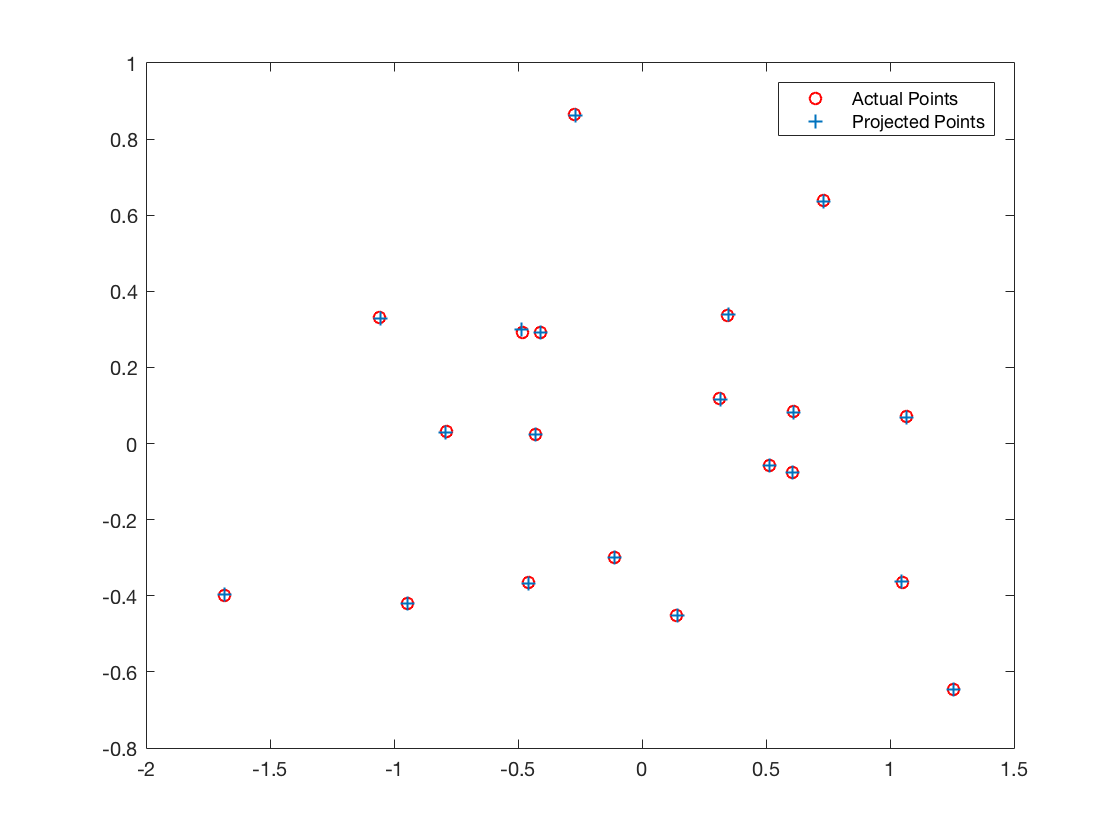
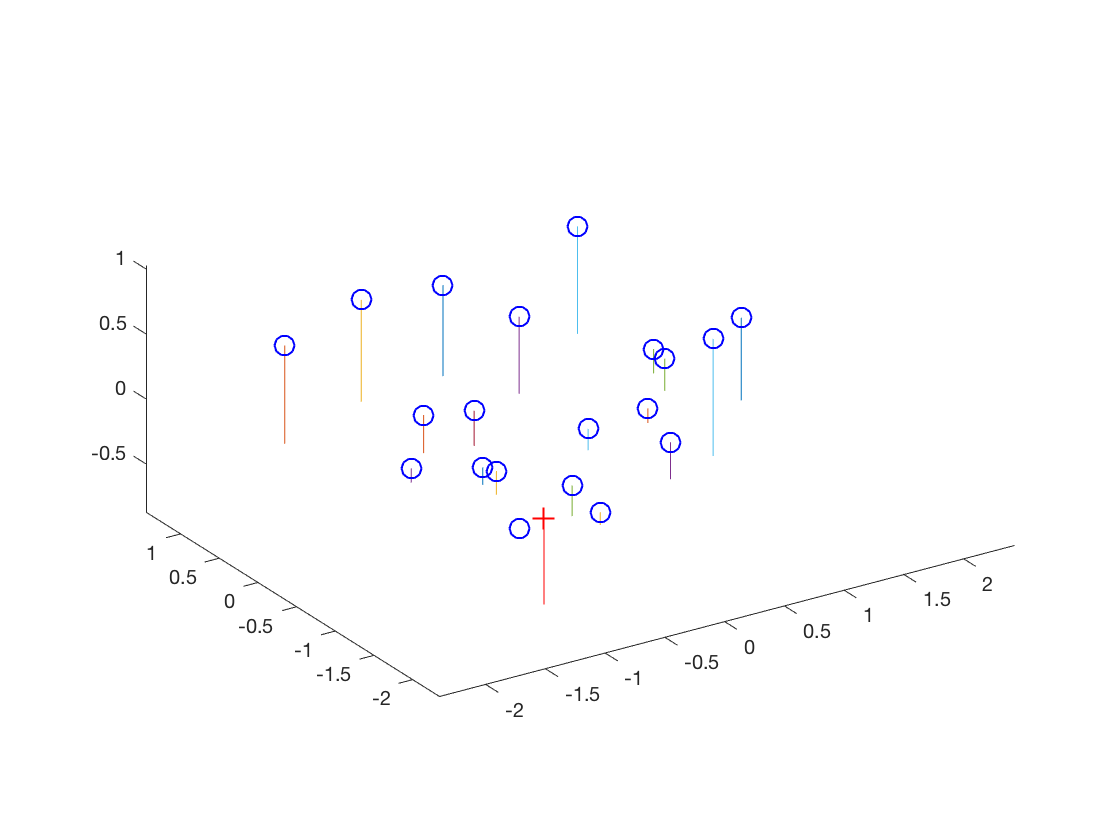
| Normalized input points | |
The projection matrix is:
0.4583 -0.2947 -0.0140 0.0040
-0.0509 -0.0546 -0.5411 -0.0524
0.1090 0.1783 -0.0443 0.5968
The total residual is: 0.0445
The estimated location of camera is: (-1.5127, -2.3517, 0.2826)
|
|
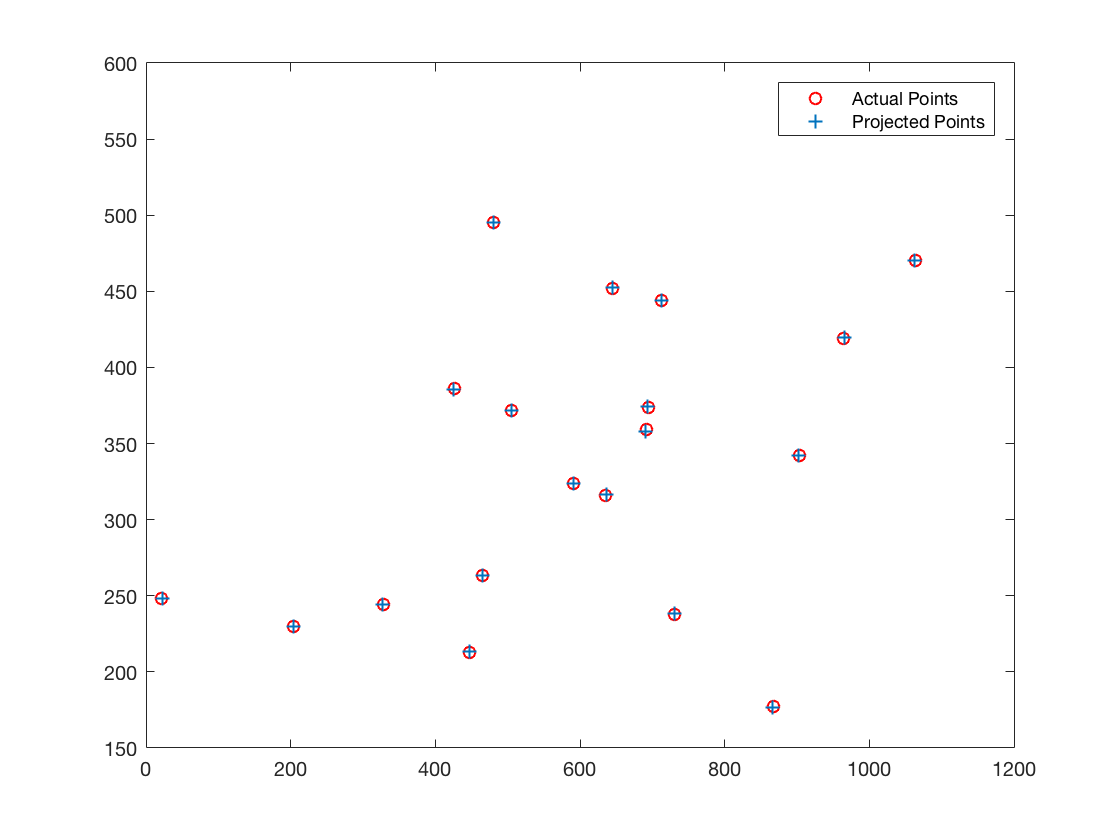
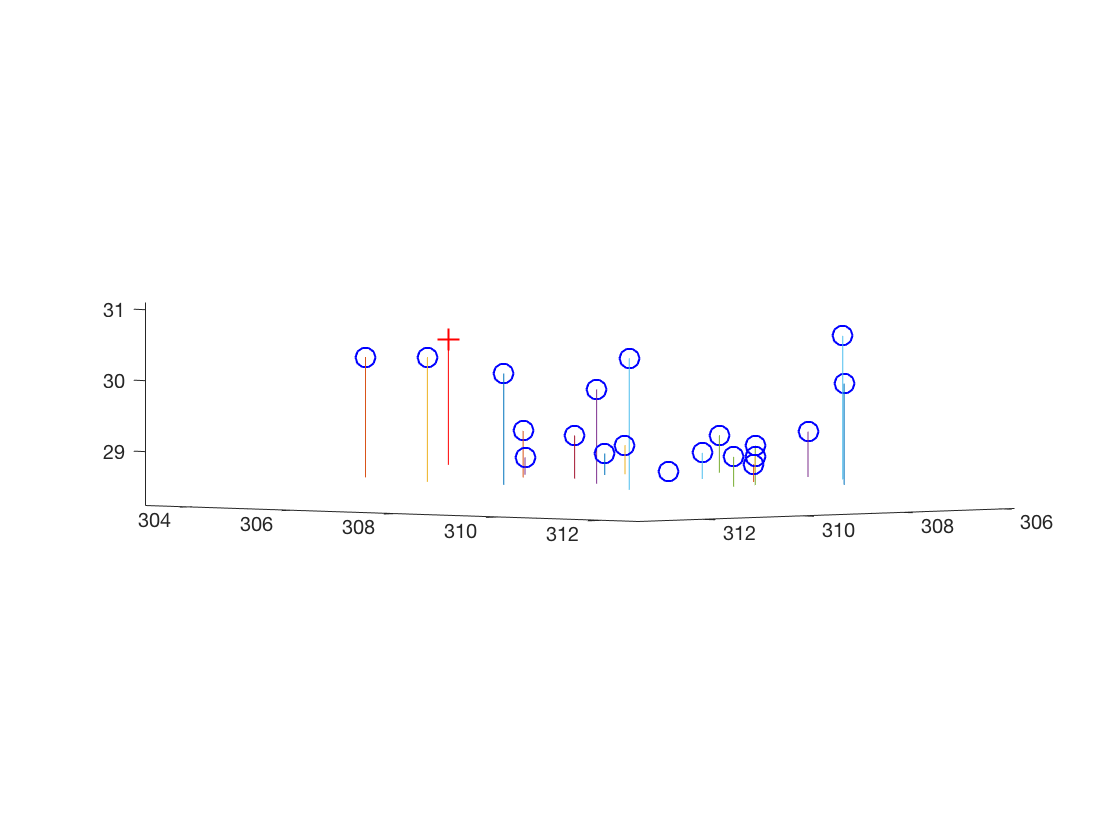
| Not-normalized input points | |
The projection matrix is:
0.0069 -0.0040 -0.0013 -0.8267
0.0015 0.0010 -0.0073 -0.5625
0.0000 0.0000 -0.0000 -0.0034
The total residual is: 15.5450
The estimated location of camera is: (303.1000, 307.1843, 30.4217)Fundamental Matrix Estimation
My
- Step 1: Setup a homogenous set of equations using the corresponding 2D and 3D points (Ax = 0)
- Step 2: Find SVD of matrix A.
- Step 3: Compute Fundamental matrix as
[U D V]= svd(A);
F = V(:,end);
F = reshape(F,[],3)';
- Step 4: Since F has rank 3, reduce its rank to 2 by computing its SVD:
[Uf Df Vf]= svd(F);
Df(end, end) = 0;
F_matrix = Uf*Df*Vf';
Extra Credit : Fundamental Matrix Estimation
To improve the estimated Fundamental Matrix, I normalized the input points. The
- Step 0.0: For both the images, find the transformation matrix T which shifts the mean of the points to origin and scales them to have a magnitude of around 1.
- Step 0.1: Now transform both the images by multiplying by T.
% Estimate scale and mean for a
mean_ax = mean(Points_a(:,1));
mean_ay = mean(Points_a(:,2));
scale_ax = 1/std(Points_a(:,1)-mean_ax );
scale_ay = 1/std(Points_a(:,2)-mean_ay );
% Transform a
Points_a = [Points_a ones(n,1)];
Points_a = (T_a*Points_a(:,1:3)')';
- Step 1: Setup a homogenous set of equations using the corresponding 2D and 3D points (Ax = 0)
- Step 2: Find SVD of matrix A.
- Step 3: Compute Fundamental matrix as
[U D V]= svd(A);
F = V(:,end);
F = reshape(F,[],3)';
- Step 4: Since F has rank 3, reduce its rank to 2 by computing its SVD:
[Uf Df Vf]= svd(F);
Df(end, end) = 0;
F_matrix = Uf*Df*Vf';
- Step 5: Now inverse transform the F obtained by:
F_matrix = T_b' * F_matrix * T_a;
Results:
|
|

|

|
| Without normalized input points: Slight difference between the lines and dots is present. | |
|
|

|

|
| With normalized input points: Almost perfect crossing of lines on dots. | |
Conclusion: Even though here not much difference is observed (because the input correspondences are pretty much perfect), we expect to see a better improvement in the next part - RANSAC.
Fundamental Matrix with RANSAC
For this Select-Compute-Update type algorithm, I tried a bunch of values for the different parameters such as:
- Number of iterations (1000, 3000, 5000): I chose 3000 as the number of inliers for 3000 and 5000 were almost the same.
- Threshold (0.1, 0.01, 0.005, 0.001): I chose 0.005 as the threshold. I preferred to achieve a
higher accuracy of inliers, although this low threshold did reduce the number of inliers.
My
- Step 1: Pick any 8 corresponding matches at random.
- Step 2: Compute the fundamental matrix as in the previous part using just these 8 points.
- Step 3: Using 0.005 as a thershold, filter out the outliers and count the number of inliers. Record this as the score of the fundamental matrix chosen.
- Step 4: Repeat the above for about 3000 iterations of RANSAC and find the best fundamental matrix with most number of inliers (max score).
RESULTS!
Below are the results for RANSAC using 0.005 as a threshold, for both versions of part 2 [un-normalised(BEFORE) and normalised(AFTER) points used for fundamental matrix estimation]
Mount Rushmore Pair Of Images (BEFORE)

|

|

| |
| Without normalized input points: About 137 inliers found. | |
Mount Rushmore Pair Of Images (AFTER)
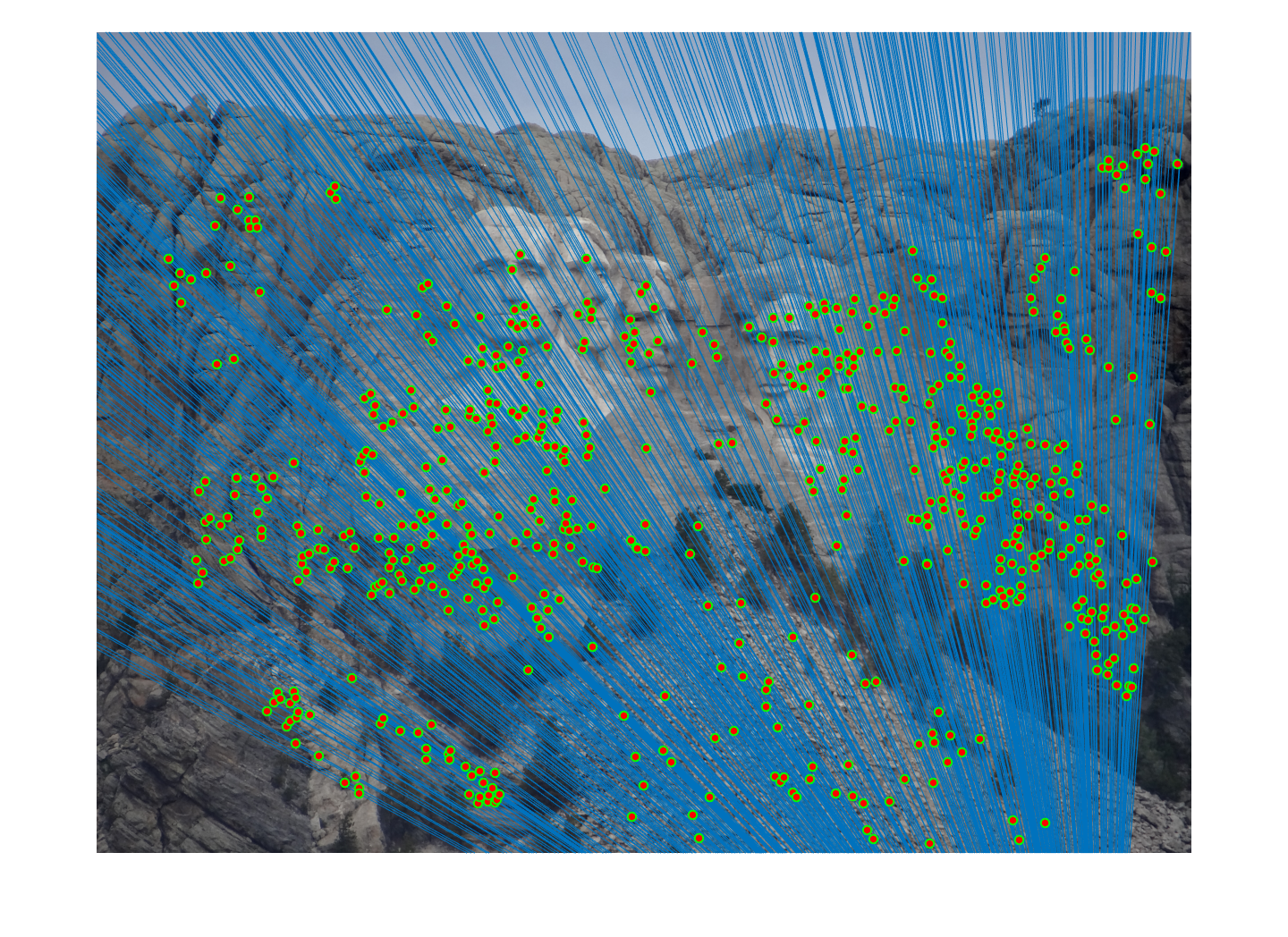
|
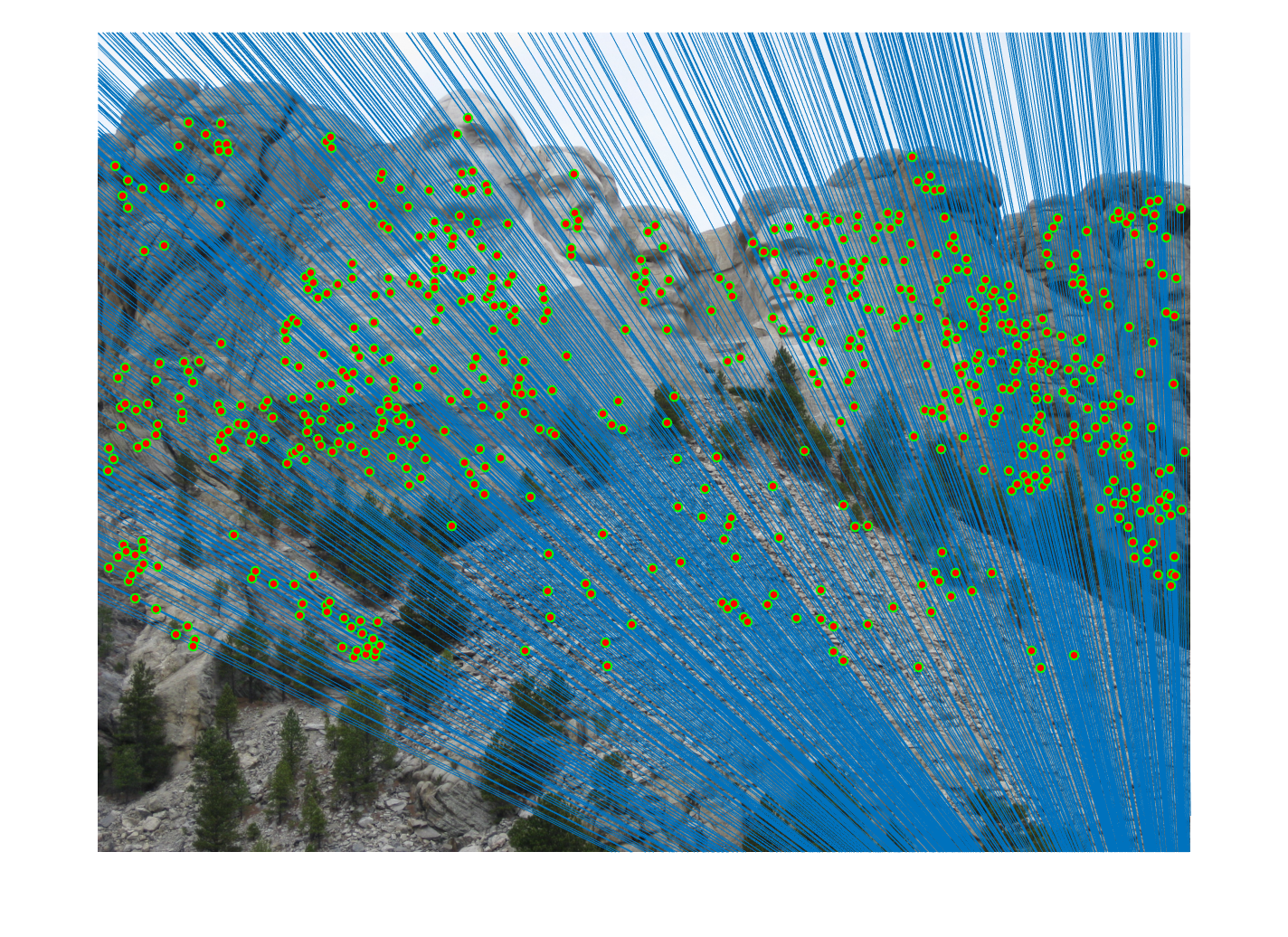
|
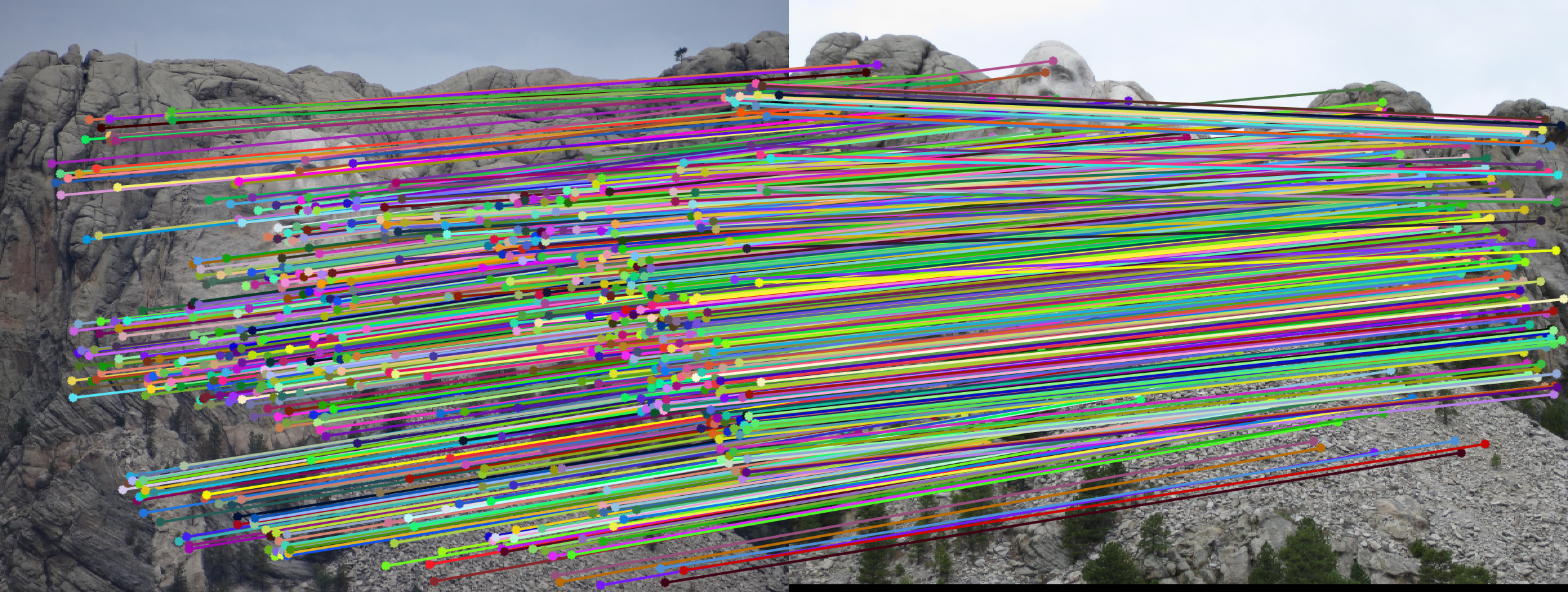
| |
| After normalization: About 649 inliers found. | |
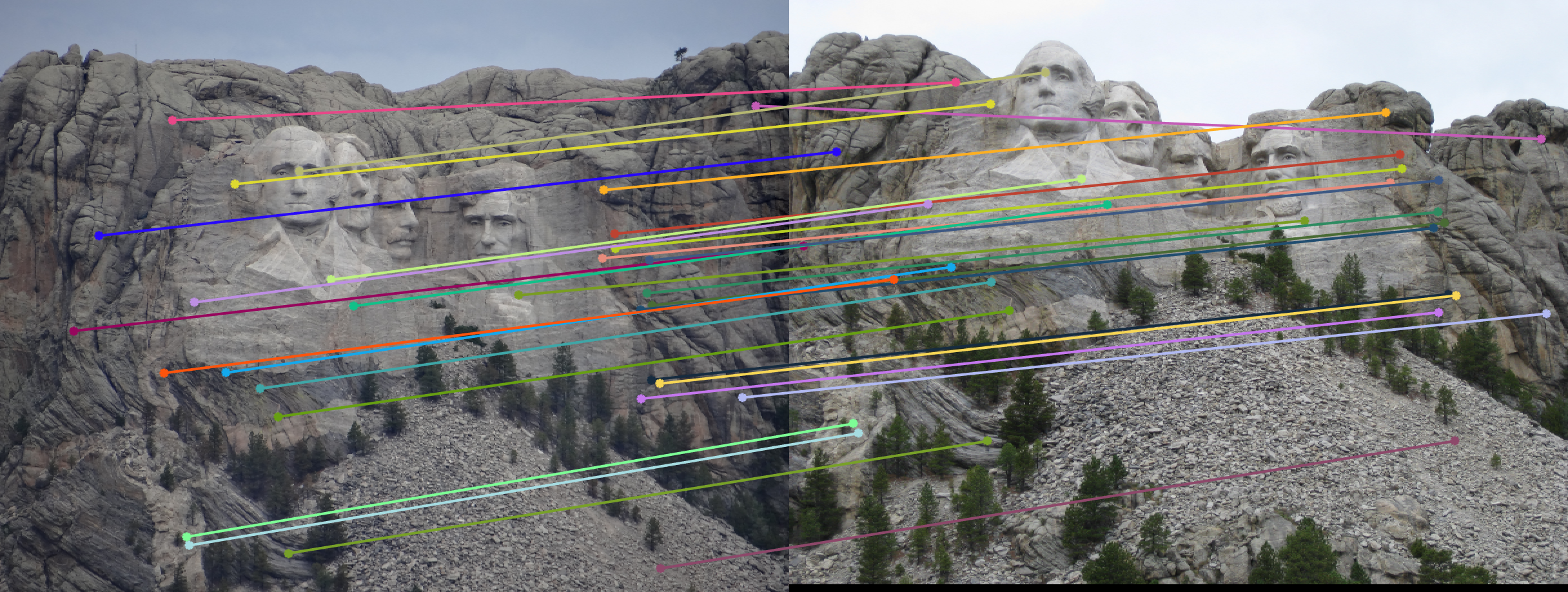
| |
| Reducing it to 30 for a neater visualization. | |
Notre Dame Pair Of Images (BEFORE)

|

|
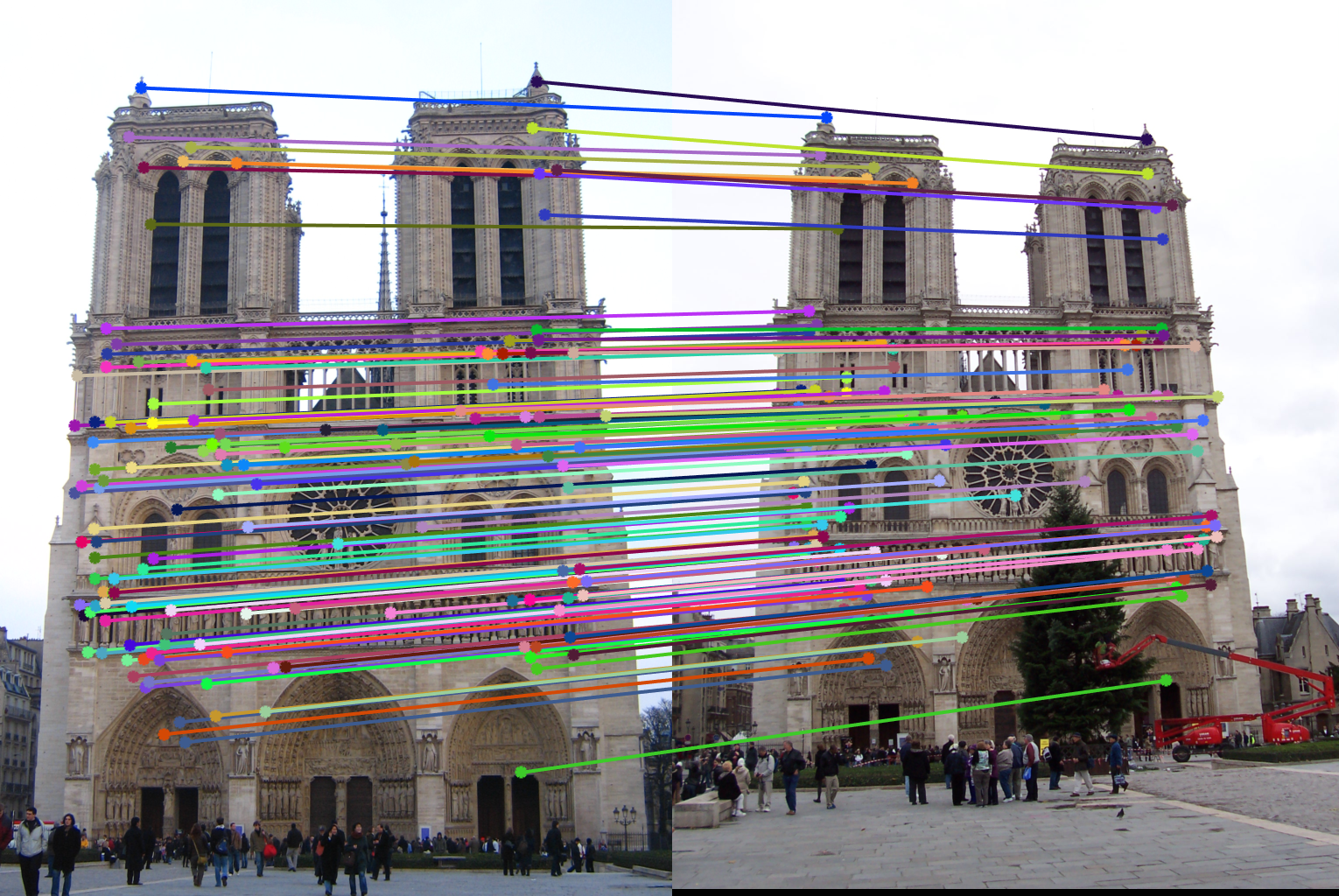
| |
| Without normalized input points: About 135 inliers found. | |
Notre Dame Pair Of Images (AFTER)

|

|

| |
| After normalization: About 520 inliers found. | |
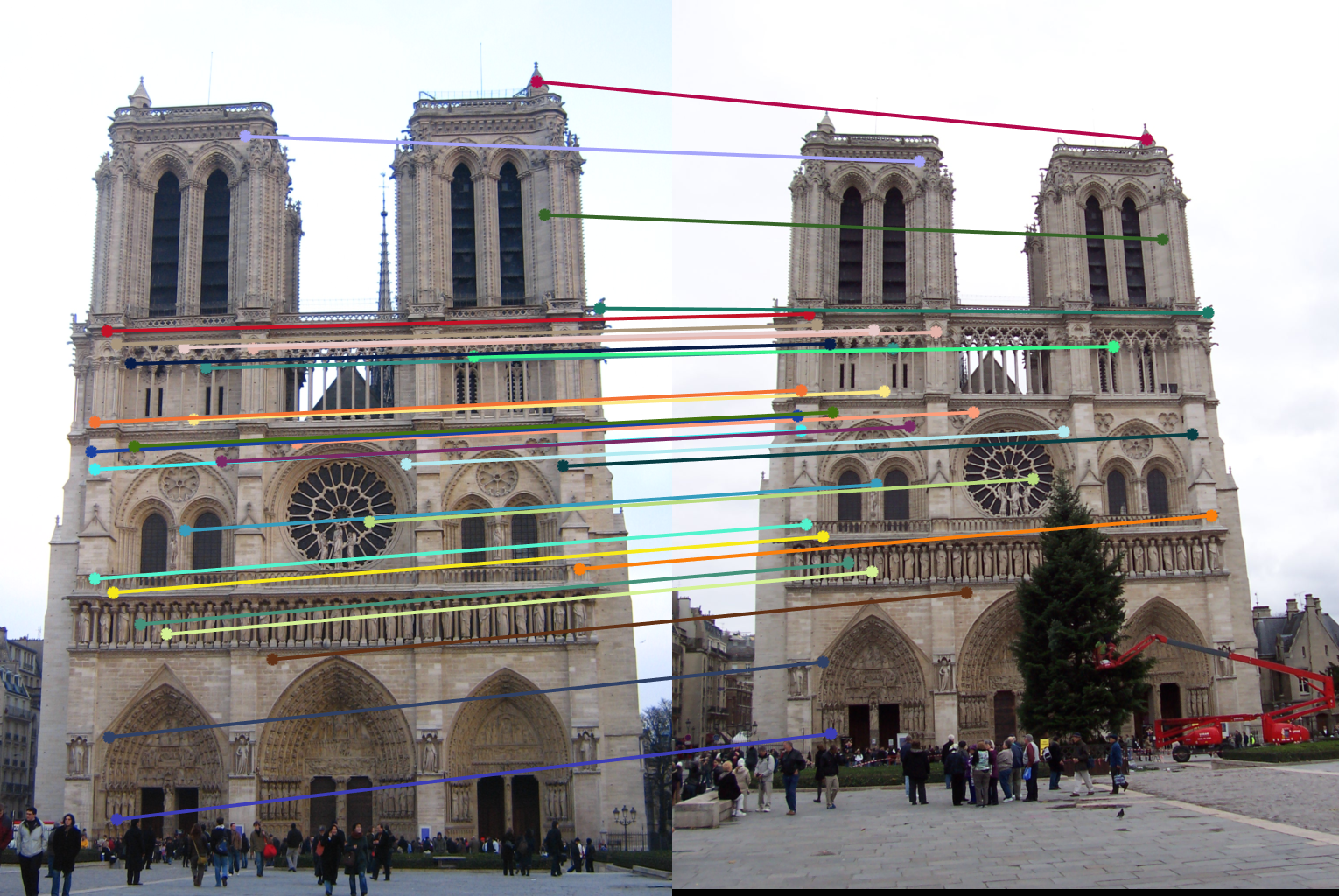
| |
| Reducing it to 30 for a neater visualization. | |
Woodruff Dorm Pair Of Images (BEFORE)

|

|

| |
| Without normalized input points: About 200 inliers found. | |
Woodruff Dorm Pair Of Images (AFTER)

|

|

| |
| After normalization: About 212 inliers found. | |

| |
| Reducing it to 30 for a neater visualization. | |
Episcopal Gaudi Pair Of Images (BEFORE)

|

|

| |
| Without normalized input points: About 202 inliers found. Note how some points are completely incorrectly matched. | |
Episcopal Gaudi Pair Of Images (AFTER)

|

|

| |
| After normalization: About 374 inliers found. Much better matched due to normalization. | |

| |
| Reducing it to 30 for a neater visualization. | |
Conclusion :
For the un-normalized points, the fundamental matrix isn't as accurate as the that for the normalized points. For a threshold of 0.005, the normalization helps achieve more inliers and more accurate inliers when RANSAC is performed to estimate the fundamental matrix.