Project 3 / Camera Calibration and Fundamental Matrix Estimation with RANSAC
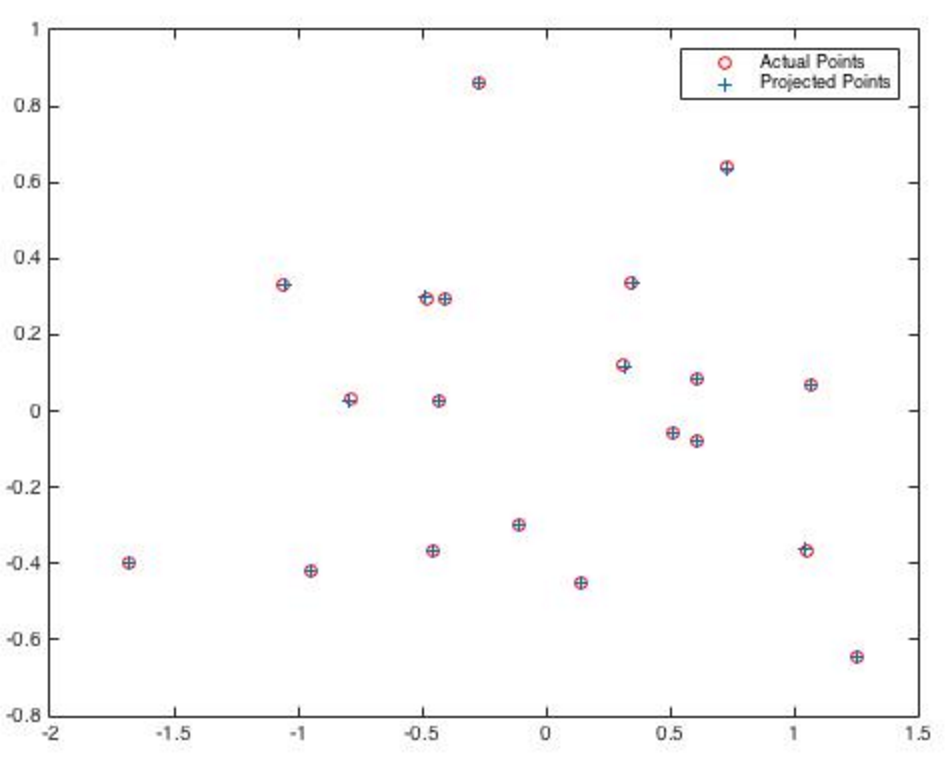
Using a projection matrix to approximate points in a 2D plane .
To the right is an example of estimating locations using a projection matrix.
The red circles are known locations in the graph used to calculate a projection matrix.
The blue pluses utilize this projection matrix to calculate these examples.
This projection matrix M is shown below.
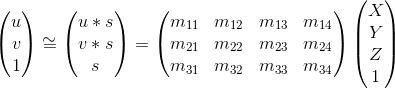
M encompasses matrix transformations including rotation and translation.
Using Singular Value Decomposition (SVD), the values of M may be solved.
This matrix may then be used to estimate project points in 2D to 3D, as well as find the location of the camera center.
An example of this is shown in the table at the end of this page.
RANSAC
Below is the process for finding a fundamental matrix F using RANSAC. F may be used to project points from one image A onto another image B. In order to evaluate any given F, we calculate the number of inliers. Corresponding points are an inlier if its projection from A into B is within a threshold of the actual point in B.
- Take a random sample of corresponding points from images A and B,
- Estimate the fundamental matrix F using this random sample,
- Project points from one image to the other using
F, - Count the number of inliers, if it is better than the best yet, record the number and this F as best.
The below steps are looped for N times or until an ideal F has been found. Once the best F has been found, we can return the most confident set of points to visualize. For an application of this process, you would most likely return all of the inliers, however we only return 30 to make the visualization clear as shown at the end of this page. The second step, estimating the fundamental matrix, boils down to solving for the matrix in the below equation, where the first vector is a coordinate in image B and the second vector is a corresponding point in image A.
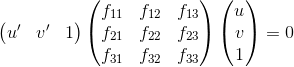
RANSAC in detail
Below is my MATLAB implemtation of RANSAC.
It heavily utilizes concurrent matrix operations to calculate the pairwise distances.
The reasoning behind overcalculating distances was to avoid the use of for loops which are notoriously slow in MATLAB.
The estimate_fundamental_matrix call utilizes SVD to compute the matrix from the given sample.
The random sample size s is some arbitrary small number and threshold is the margin of error we allow to count a point as an inlier.
There are several hyperparameters which may be adjusted to tune performance.
The probability p is the probability of finding a bad estimation with our random sample size.
The outlier_ratio is a measure of the ratio of number of outliers to total points.
These values are chosen for ideal performance.
Smaller samples allow for the fundamental matrix to be estimated quickly and leads to less iterations in N.
Lower probabilities lead to less iterations which may result in a poorly performing fundamental matrix.
Likewise, lower outlier_ratios lead to less iterations, something which may not be advantageous.
The inlier_ratio is similar to outlier_ratio however it is the ratio of inliers to total points.
%RANSAC
function [ Best_Fmatrix, inliers_a, inliers_b] = ransac_fundamental_matrix(matches_a, matches_b)
num_matches = min(size(matches_a, 1), size(matches_b, 1));
matches_a_3d = [matches_a, ones(num_matches, 1)];
matches_b_3d = [matches_b, ones(num_matches, 1)];
s = 8;
p = 0.99;
threshold = 0.05;
outlier_ratio = 0.5;
inlier_ratio = 0;
N = log(1 - p) / log(1 - (1 - outlier_ratio) ^ s);
for i=1:N
indices = randsample(num_matches, s);
sample_a = matches_a(indices, :);
sample_b = matches_b(indices, :);
F_matrix = estimate_fundamental_matrix(sample_a, sample_b);
distances = diag(abs(matches_b_3d * F_matrix * matches_a_3d'));
num_inliers = size(find(distances < threshold), 1);
if (num_inliers / num_matches) > inlier_ratio
inlier_ratio = num_inliers / num_matches;
Best_Fmatrix = F_matrix;
end
end
distances = diag(abs(matches_b_3d * Best_Fmatrix * matches_a_3d'));
[~, indices] = sort(distances);
% either return all points or the number of inliers, whichever is smaller
num = min(num_matches, floor(inlier_ratio * num_matches);
% reindex using sorted indices
inliers_a = matches_a(indices, :);
inliers_b = matches_b(indices, :);
% return only the top num corresponding points
inliers_a = inliers_a(1:num, :);
inliers_b = inliers_b(1:num, :);
Results in a table
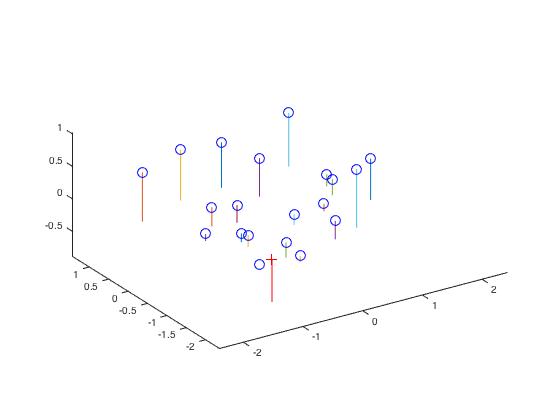
|
| Estimating the camera center using a 2D to 3D projection matrix. The blue circles are the points and the red plus is the estimated camera center. |


|
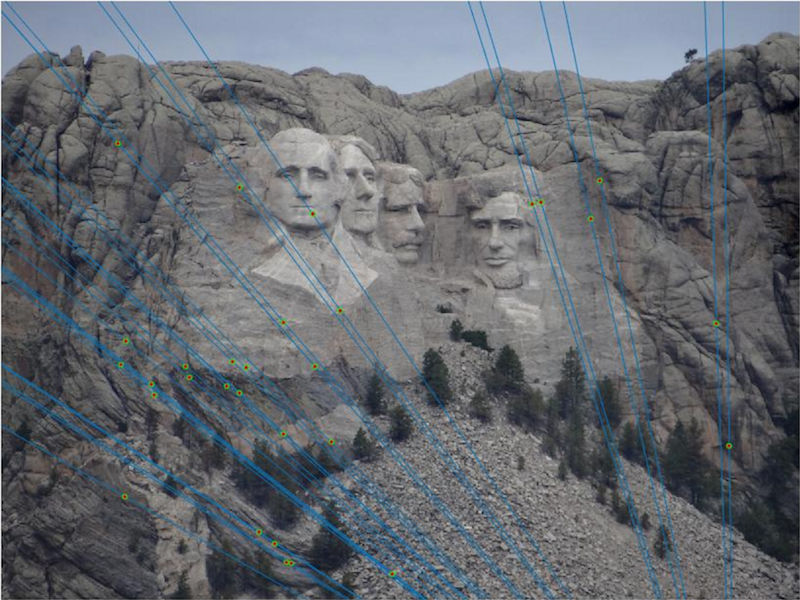
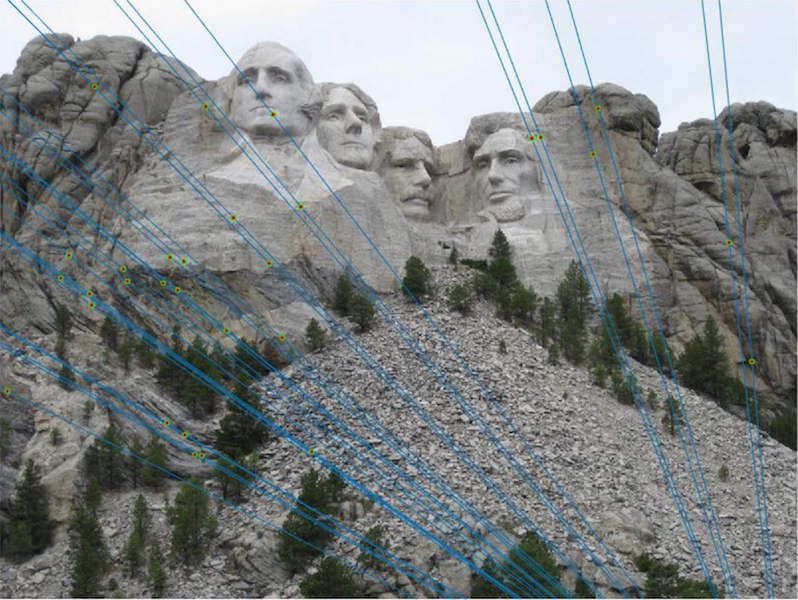
| Two images taken of the same scene from different camera centers. The epipolar lines are overlayed using an estimated fundamental matrix. The location of the markers is known, and each line approximately intersects the known locations. |


|
| The original left and right images. |

|
| Matching points using a SIFT pipeline. displaing several incorrect matches evident. There are approximately 800 corresponding points in this image. |
|
| The 30 most confident epipolar lines drawn on top of the original images. |

|
| The 30 most confident corresponding points found using RANSAC. |

|
| All corresponding points considered inliers found using RANSAC. There are approximately 500 pairs in this image. |
|
|
| Using RANSAC to match points on Notre Dame. This case is challening since most points are in the same plane. |
|
|
| Using RANSAC to match points on Episcopal Gaudi. The accuracy for this case is significantly less than Mount Rushmore or Notre Dame. |
The RANSAC results in the above table all used the same parameters shown in the RANSAC in detail section.
Tests took approximately 15 seconds to run, a good compromise between accuracy and speed.
Both Mount Rushmore and Notre Dame performed very well using RANSAC, however Episcopal Gaudi had some evident accuracy problems.
One possible improvement to the RANSAC pipeline is in the estimate_fundamental_matrix function.
By normalizing the passed in coordinates before estimating the fundamental matrix.
This modification has been suggested to improve results across the board without a noticeable impact to speed.