Project 3 / Camera Calibration and Fundamental Matrix Estimation with RANSAC
This project is designed to build on top of project 2 in terms of improving the feature matching process between two images. The project is broken down into 2 parts: calculating the camera projection matrix (extrinsic) and camera center, and using the RANSAC algorithm to estimate the optimal fundamental matrix
Camera Projection Matrix and Center
The projection matrix formula is (x,y,1) = K*[R T]*(u,v,w,1)^T whereby (u,v,w,1) are homogeneous coordinates.
- We are computing the projection matrix by constructing a linear system such that Ax = 0
- Next we are using Singular Value Decomposition to minimize the residual value
- I used a scale of -1 for my projection matrix to match the values given for the assignment.
- To solve for the camera center, I used the formula Center = -Q^-1*M_4, whereby Q is M(1:3) and is a square matrix
My projection matrix
My Camera Center
My residual
0.0445
Finding Best Fundamental Matrix with RANSAC
Algorithm for Fundamental Matrix Estimation
- Since I did the extra credit for this assignment, I first created the Transform matrices T_a and T_b by using scale s of 1/abs(mean_u - mean_v) to create the offset and scale matrices for both A and B
- For each point I normalized the point using the aforementioned Transform matrices, and then created a linear system
- I ran SVD on the linear system and obtained F from matrix V. However, since the fundamental matrix is of rank 2, I ran SVD again after zeroing out the first value in the diagonal matrix S.
- Finally F_matrix = U*S*V' whereby F_matrix is of rank 2.
Obtaining optimal fundamental Matrix
- We are going to be exploiting the fact that x'T*F*x = 0 whereby x' and x are two camera locations. However, since F is approximate, we will testing for x'T*F*x < threshold
- I set the maximum number of iterations to 2000 and the threshold of the aforementioned equation to 0.02
- To implement RANSAC, I sampled out 8 points using the randsample function from n points and obtained inliers using the aforementioned formula.
- To ensure that I obtain the best fundamental matrix, I reassign the inlier_points to the F that maximizes the number of inliers.
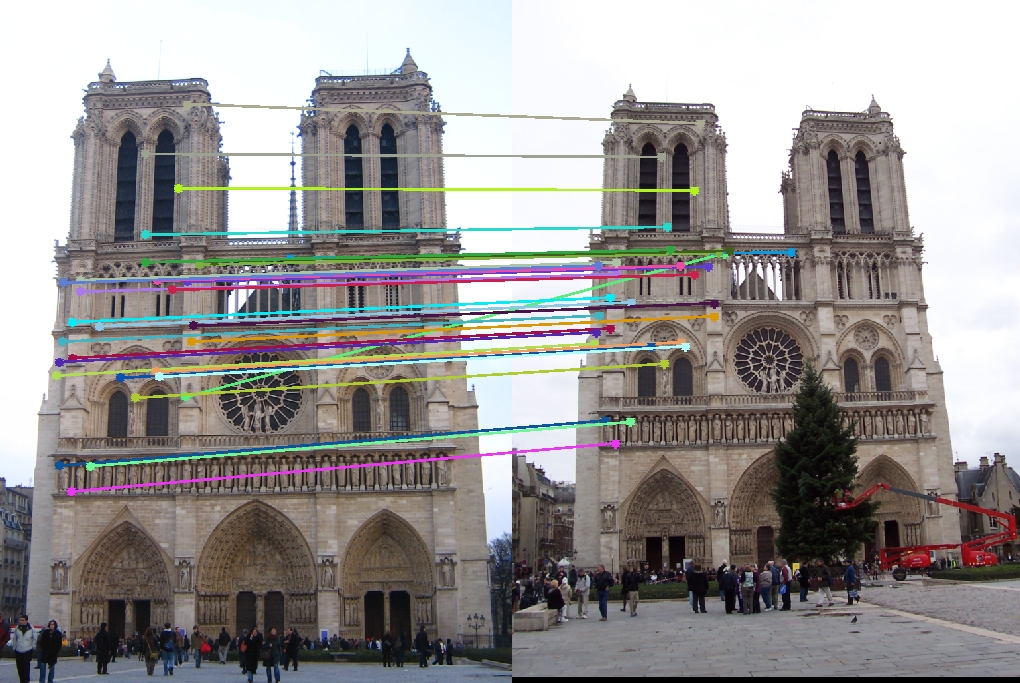
Results with normalization
The following results display 30 random inliers. All of the image pairs show only around 1 or 2 inliers that are incorrectly marked
| Vis Arrows | Left Epipolar Image | Right Epipolar Image | Best Fundamental Matrix |
|---|---|---|---|
 |
 |
 |
|
 |
 |
 |
|
 |
 |
 |
|
 |
 |
 |
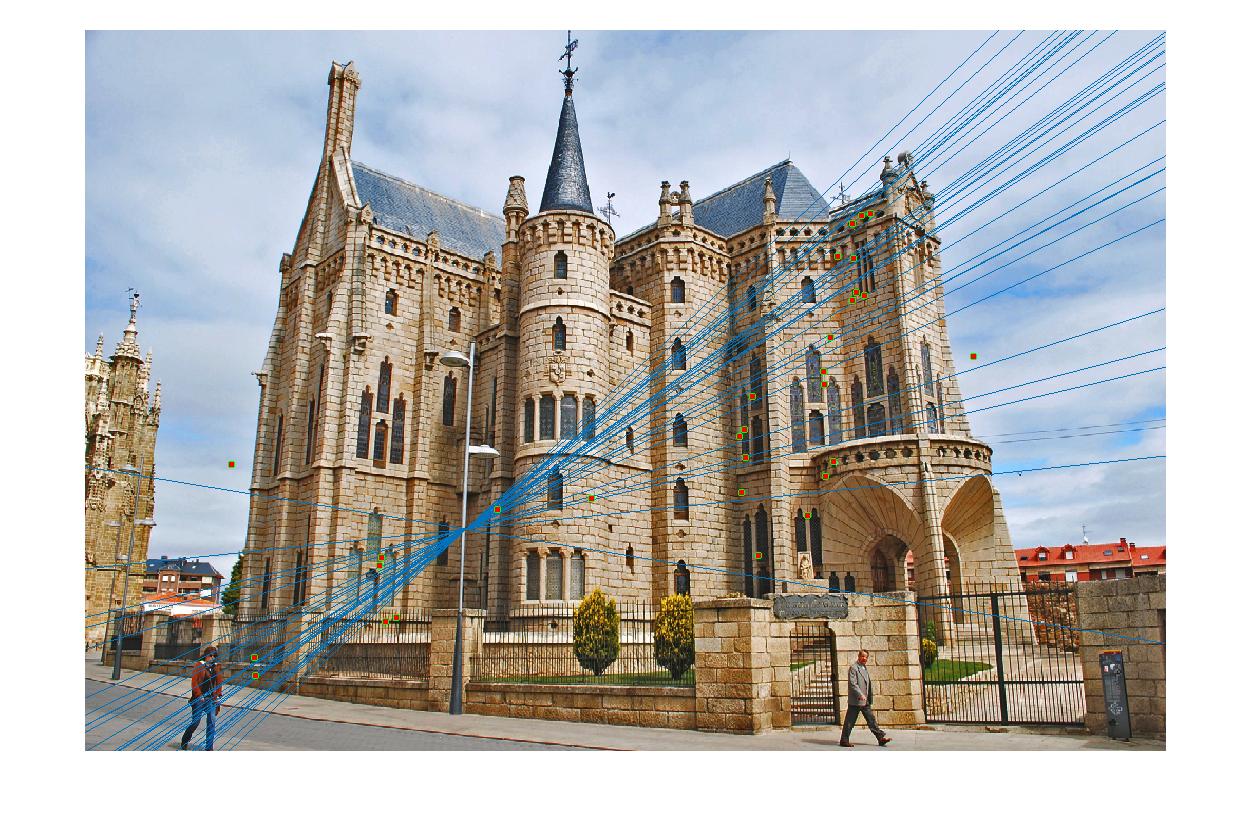
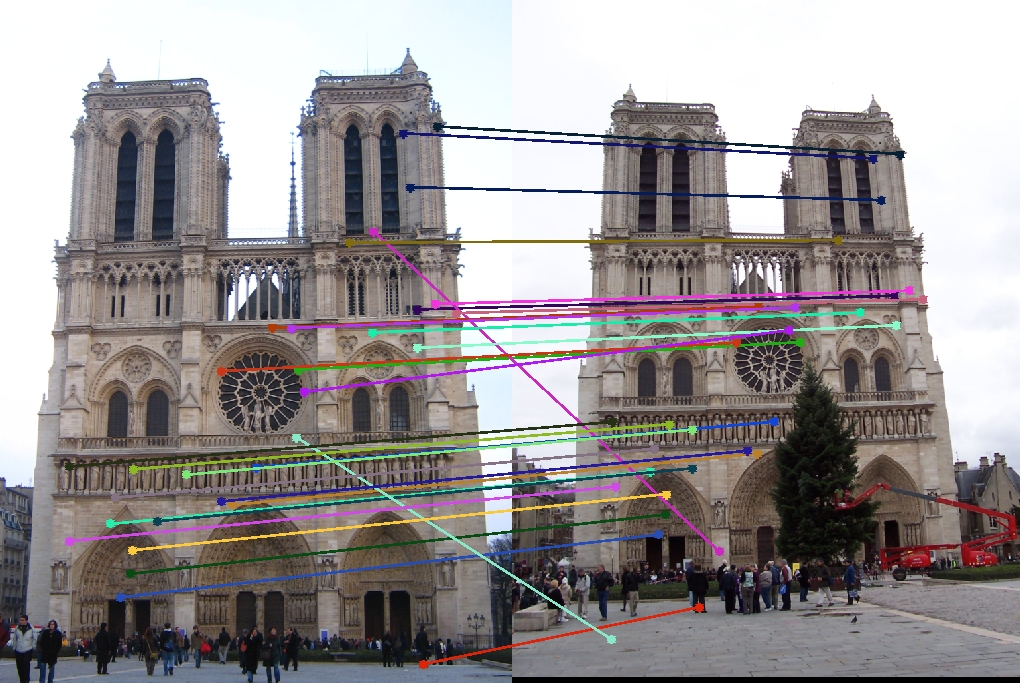
Results without normalization
I'm using Gaudi and Notre Dame to demonstrate the effectiveness of normalization. As shown here, the accuracy of 30 random inliers for Gaudi and Notre Dame without normalization is <= 76.67%
| Vis Arrows | Left Epipolar Image | Right Epipolar Image | Best Fundamental Matrix |
|---|---|---|---|
 |
 |
 |
|
 |
 |
 |
Extra Credit
The two tables above demonstrate the effect of coordinate normalization and that the accuracy is boosted with its implementation.
Implementation of extra credit
n = length(Points_a);
A = zeros(n, 9);
mean_a_u = mean2(Points_a(:,1));
mean_a_v = mean2(Points_a(:,2));
mean_b_u = mean2(Points_b(:,1));
mean_b_v = mean2(Points_b(:,2));
scale_a = 1/(abs(mean_a_u - mean_a_v));
scale_b = 1/(abs(mean_b_u - mean_b_v));
scale_matrix_a = [scale_a 0 0;...
0 scale_a 0; ...
0 0 1];
scale_matrix_b = [scale_b 0 0;...
0 scale_b 0; ...
0 0 1];
offset_matrix_a = [1 0 -mean_a_u;...
0 1 -mean_a_v; ...
0 0 1];
offset_matrix_b = [1 0 -mean_b_u;...
0 1 -mean_b_v; ...
0 0 1];
T_a = scale_matrix_a*offset_matrix_a;
T_b = scale_matrix_b*offset_matrix_b;
for i=1:n
a = T_a*[Points_a(i,:) 1]';
b = T_b*[Points_b(i,:) 1]';
a1 = a(1,1);
a2 = a(2,1);
b1 = b(1,1);
b2 = b(2,1);
A(i, :) = [b1*a1 b1*a2 b1 b2*a1 b2*a2 b2 a1 a2 1];
end
[U, S, V] = svd(A);
F = V(:,end);
F = reshape(F, [3 3])';
[U, S, V] = svd(F);
S(3,3) = 0;
F_matrix = U*S*V';
F_matrix = T_b'*F_matrix*T_a;
end