Project 3 / Camera Calibration and Fundamental Matrix Estimation with RANSAC

Example of a right floating element.
This project was experimenting camera projection matrix and fundamental matrix. This assignment had three parts:
- Camera Projection Matrix
- Fundamental Matrix Estimation
- Fundamental Matrix with RANSAC
Part I: Camera Projection Matrix
This first part was calculating the projection matrix given a set of 2D and 3D points. This was done by using method 1 in lecture 13 slides. This method calculates the m matrix values by using the V matrix from SVD. Taking a column vector from here gives us a potential M Matrix. We then calculate the camera's world coordinates by working with [R|T] Matrix.
Part 1 results
The projection matrix is:
-0.4583 0.2947 0.0140 -0.0040
0.0509 0.0546 0.5411 0.0524
-0.1090 -0.1783 0.0443 -0.5968
The total residual is: <0.0445>
The estimated location of camera is: <-1.5127, -2.3517, 0.2826>
Part 1 Code
function M = calculate_projection_matrix( Points_2D, Points_3D )
numPoints = size(Points_2D,1);
Y = reshape(Points_2D', 2*numPoints, []);
A = zeros(2*numPoints, 12);
for row = 1:numPoints
curr3D = Points_3D(row,:);
A(2*row-1,:) = [curr3D, 1, zeros(1,4), -Y(2*row-1)*curr3D, -Y(2*row-1)];
A(2*row,:) = [zeros(1,4), curr3D, 1, -Y(2*row)*curr3D, -Y(2*row)];
end
[U, S, V] = svd(A);
M = V(:,end);
M = -reshape(M,[],3)`;
end
function [ Center ] = compute_camera_center( M )
M4 = M(1:3, 4);
Q = -inv(M(1:3, 1:3));
Center = Q * M4;
end
Part II: Fundamental Matrix Estimation
We are given a collection of 2D/3D corresponding points. We find the fundamental matrix by rearranging the Fundamental Matrix definition to create a linear system of equations. We solve the homogeneous linear equation by using the 8-point algorithm. We use the SVD idea again. We change the Fundamental matrix by calculating SVD and setting (3,3) to 0 because of the matrix being rank 2.
Part 2 Results
Fundamental Matrix Estimation:
| -1.97289236987104e-07 2.70940307021885e-06 -0.000677036142197256 |
| 1.87658315555846e-06 -4.62330421606139e-07 0.00545515857531132 |
| -4.05830999075855e-05 -0.00749544531661520 0.174040454346722 |
Extra Credit
I included extra credit by normalizing the coordinates before computing the Fundamental Matrix. I find the mean coordinates for each image, center the images around the origin, and then calculate the standard deviation to create two transform matricies. At the end of calculating the original fundamental matrix, I apply the transformations again to scale to the original image. This improved my images by centering every line on the keypoints whereas without it some of the lines were off point.
Part 2 Code
function [ F_matrix ] = estimate_fundamental_matrix(Points_a,Points_b)
numPoints = size(Points_a, 1);
A = zeros(numPoints, 9);
c_u = sum(Points_a(:,1))/numPoints;
c_v = sum(Points_a(:,2))/numPoints;
c_up = sum(Points_b(:,1))/numPoints;
c_vp = sum(Points_b(:,2))/numPoints;
offset_a = [1 0 -c_u; 0 1 -c_v; 0 0 1];
offset_b = [1 0 -c_up; 0 1 -c_vp; 0 0 1];
for i = 1:numPoints
Points_a(i, :) = Points_a(i, :) - [c_u, c_v];
Points_b(i, :) = Points_b(i, :) - [c_up, c_vp];
end
s_u = 1/std(Points_a(:,1));
s_v = 1/std(Points_a(:,2));
s_up = 1/std(Points_b(:,1));
s_vp = 1/std(Points_b(:,2));
scale_a = [s_u 0 0; 0 s_v 0; 0 0 1];
scale_b = [s_up 0 0; 0 s_vp 0; 0 0 1];
for i = 1:numPoints
Points_a(i, :) = diag([s_u, s_v]) * Points_a(i, :)';
Points_b(i, :) = diag([s_up, s_vp]) * Points_b(i, :)';
end
T_a = scale_a * offset_a;
T_b = scale_b * offset_b;
for row = 1:numPoints
a = num2cell(Points_a(row, :));
b = num2cell(Points_b(row, :));
[u, v] = a{:};
[up, vp] = b{:};
A(row,:) = [u*up, u*vp, u, v*up, v*vp, v, up, vp, 1];
end
[U, S, V] = svd(A);
f = V(:, end);
F = reshape(f, [3 3]);
[U, S, V] = svd(F);
S(3,3) = 0;
F_matrix = U*S*V';
F_matrix = T_b' * F_matrix * T_a;
end
Part 2 Table: Un-normalized vs. Normalized


|


|
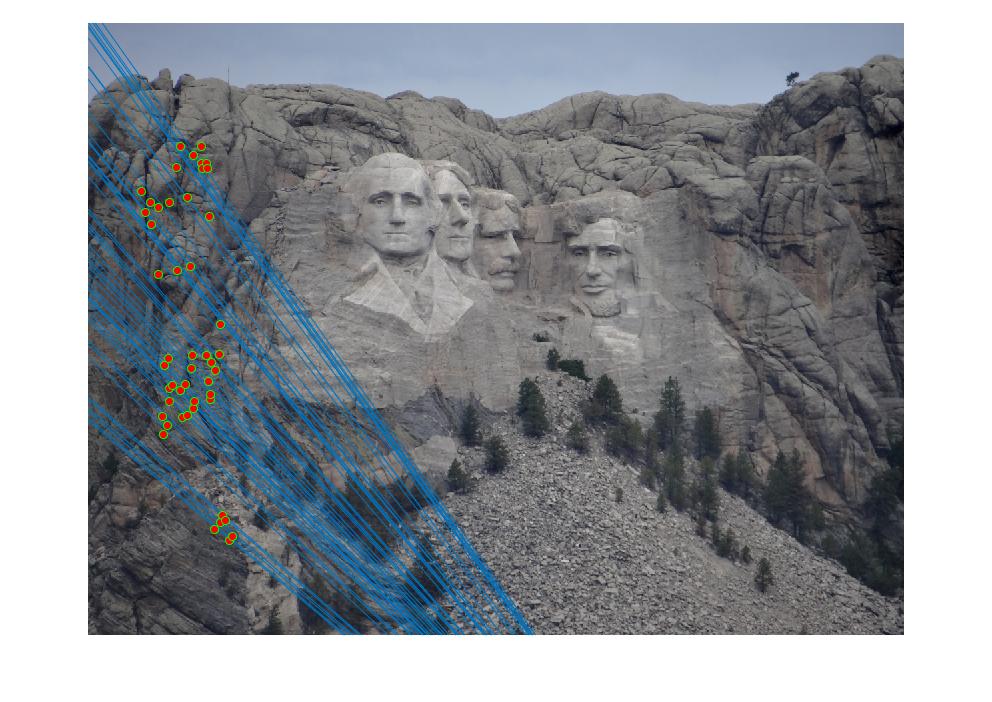
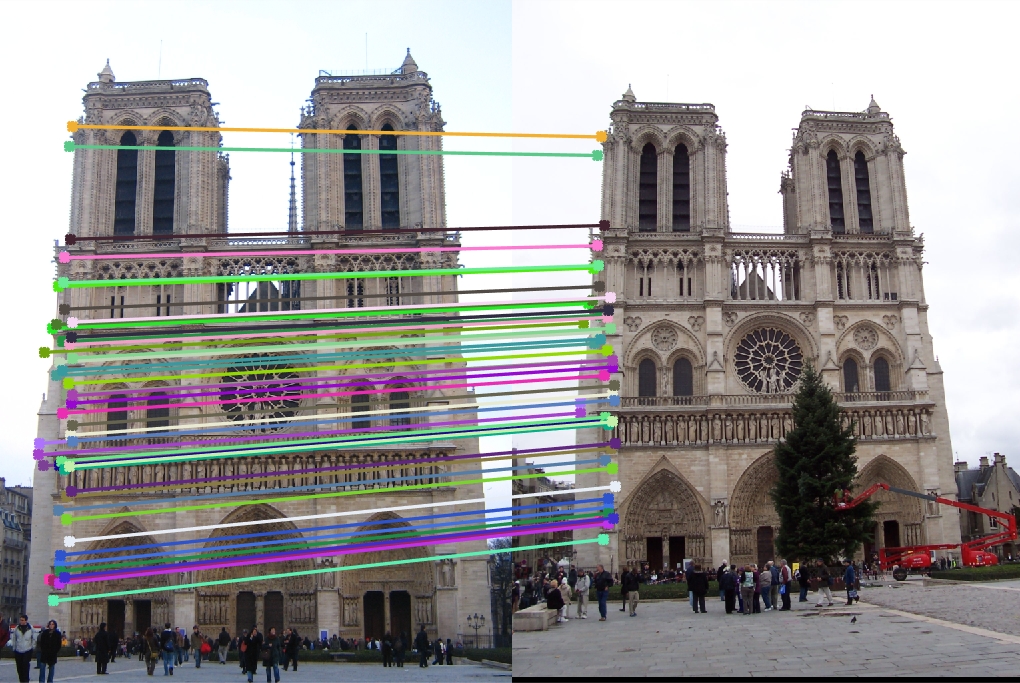
Part III: Fundamental Matrix with RANSAC
We're given two images without corresponding keypoints, so we use SIFT to find interest points. We then perform RANSAC with an iteration of 2000. We randomly sample 8 correspoinding interest points on each picture and calculate the Fundamental matrix from those points. We then go through every point and calculate an error using the definition of the Fundamental matrix (point_b * F * point_a' = 0). We use a threshold of 0.5 through trial and error (high threshold allows for too many uncorresponding points, low threshold includes too little points), and if a corresponding pair of points is within the threshold, we increment a counter. With the iterations, we find which Fundamental Matrix included the most corresponding points. We then pick only 50 points.
Part 3 Code
function [ Best_Fmatrix, inliers_a, inliers_b] = ransac_fundamental_matrix(matches_a, matches_b)
numPoints = size(matches_a,1);
inlier_threshold = 0.05;
best_inlier_count = 0;
Best_Fmatrix = [];
inliers_a = [];
inliers_b = [];
for i = 1:2000
points = randperm(numPoints, 8);
a = matches_a(points, :);
b = matches_b(points, :);
temp_F_Matrix = estimate_fundamental_matrix(a, b);
temp_count = 0;
temp_indices = [];
for j = 1:size(matches_a,1)
threshold = [matches_b(j,:) 1] * temp_F_Matrix * [matches_a(j,:) 1]';
if (abs(threshold) < inlier_threshold)
temp_count = temp_count + 1;
temp_indices = [temp_indices, j];
end
end
if temp_count > best_inlier_count
best_inlier_count = temp_count;
inliers_a = matches_a(temp_indices, :);
inliers_b = matches_b(temp_indices, :);
Best_Fmatrix = temp_F_Matrix;
end
end
inliers_a = inliers_a([1:50],:)
inliers_b = inliers_b([1:50],:)
end
Part 3 Results


|


|



|