Project 3 / Camera Calibration and Fundamental Matrix Estimation with RANSAC
Project 3 was about camera calibration and fundamental matrix estimation with RANSAC. There were three main parts to this project.
- Part 1: Camera Calibration
- Part 2: Estimating the Fundamental Matrix
- Part 3: Estimating the Fundamental Matrix with RANSAC
Part 1:
Part 1 was about finding the camera projection matrix that goes from 3D coordinates to 2D coordinates. Knowing the equation for homogeneous coordinates equation for finding the projection matrix, I solved for the projection matrix using SVD. Then I found the camera's center by finding the negative inverse of Q multiplied by the last column of M.
Calculating the Projection Matrix
function M = calculate_projection_matrix( Points_2D, Points_3D )
[rows, ~] = size(Points_2D);
A = zeros(2 * rows, 12);
for i = 1:rows
u = Points_2D(i, 1);
v = Points_2D(i, 2);
x = Points_3D(i, 1);
y = Points_3D(i, 2);
z = Points_3D(i, 3);
row1 = [x y z 1 0 0 0 0 -u*x -u*y -u*z -u];
row2 = [0 0 0 0 x y z 1 -v*x -v*y -v*z -v];
A(2 * i - 1, :) = row1;
A(2 * i, :) = row2;
end
[~, ~, V] = svd(A);
M = V(:, end);
M = reshape(M, [], 3)';
end
function [ Center ] = compute_camera_center(M)
Q = M(:,1:3);
m_4 = M(:,4);
Center = -inv(Q) * m_4;
end
Part 2:
Part 2 was about estimating the fundamental matrix. I did this by using the definition of the fundamental matrix and solving for it using SVD again like in Part 1. Unlike the first time, I used SVD again on the matrix to get a rank 2 matrix since I had a full rank matrix.
Estimating the Fundamental Matrix
function [ F_matrix ] = estimate_fundamental_matrix(Points_a,Points_b)
[n, ~] = size(Points_a);
A = zeros(n, 9);
for i = 1:n
u_a = Points_b(i, 1);
v_a = Points_b(i, 2);
u_b = Points_a(i, 1);
v_b = Points_a(i, 2);
A(i, :) = [u_b*u_a v_b*u_a u_a u_b*v_a v_b*v_a v_a u_b v_b 1];
end
[~, ~, V] = svd(A);
F = V(:, end);
F_matrix = reshape(F, [3 3])';
[U, S, V] = svd(F_matrix);
S(3, 3) = 0;
F_matrix = U * S * V';
end
Part 3:
Part 3 was about estimating the fundamental matrix with RANSAC. I first set the number of matches per sample to 8 since it is not too big or small of a number. I also set the thresh, the ratio for the number of matches inside delta to total number, to .9. I set the number of iterations to 1000 to ensure that I get a good result. I basically chose 8 indices and found 8 matches for a and b and used Part 2's code to find the fundamental matrix estimate. I used that to score the matrix and if the ratio of matches to total was greater than the thresh, then maximums for a and b would be set and end the loop since a good result was found. But if not, then it would set the new maxes if the number of matches was greater than the previous max. The score was determined by using delta = .05 and seeing if xFx' < delta.
Estimating the Fundamental Matrix with RANSAC
function [Best_Fmatrix, inliers_a, inliers_b] = ransac_fundamental_matrix(matches_a, matches_b)
thresh = 0.9;
it = 1000;
[n, ~] = size(matches_a);
max = 0;
max_a = 0;
max_b = 0;
for i = 1:it
indices = randsample(n, 8);
sample_a = matches_a(indices, :);
sample_b = matches_b(indices, :);
F = estimate_fundamental_matrix(sample_a, sample_b);
[in_a, in_b] = score_helper(matches_a, matches_b, F, n);
[inliers, ~] = size(in_a);
if inliers / n > thresh
max_a = in_a;
max_b = in_b;
break;
elseif inliers > max
max = inliers;
max_a = in_a;
max_b = in_b;
end
end
inliers_a = max_a;
inliers_b = max_b;
Best_Fmatrix = estimate_fundamental_matrix(in_a, in_b);
end
function [in_a, in_b] = score_helper(matches_a, matches_b, F, n)
in_a = [];
in_b = [];
delta = .05;
for i = 1:n
if abs([matches_b(i, :) 1] * F * [matches_a(i, :) 1]') < delta
in_a = [in_a; matches_a(i, :)];
in_b = [in_b; matches_b(i, :)];
end
end
end
Results
Results for Part 1:
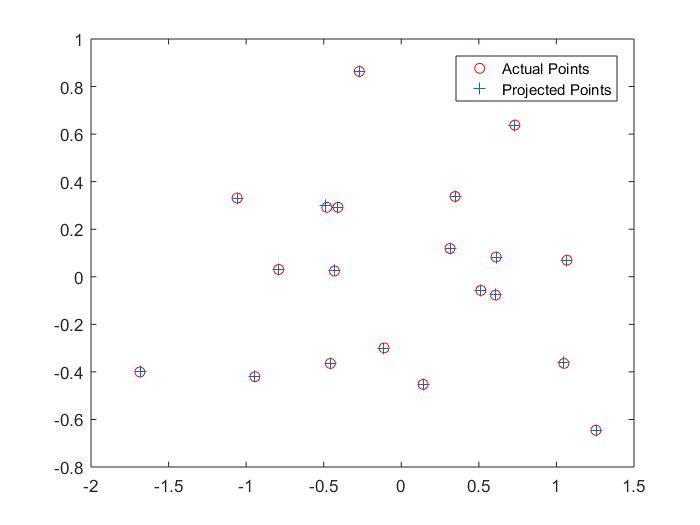
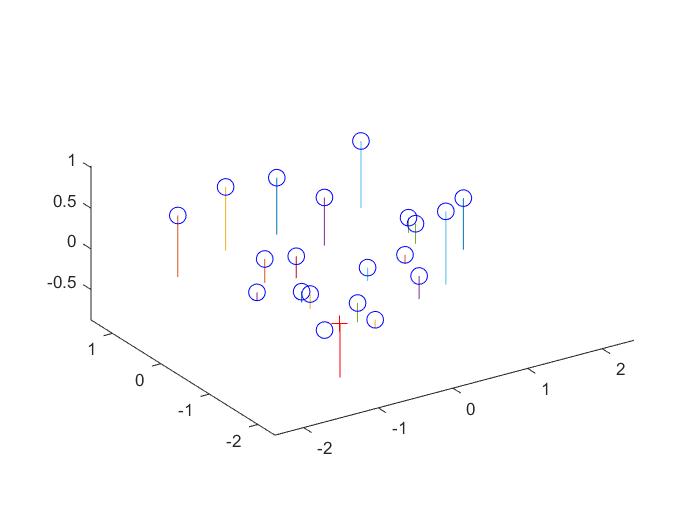
|
The projection matrix is:
0.4583 -0.2947 -0.0140 0.0040
-0.0509 -0.0546 -0.5411 -0.0524
0.1090 0.1783 -0.0443 0.5968
The total residual is: <0.0445>
The estimated location of camera is: <-1.5127, -2.3517, 0.2826>
These results are approximately right. The projection matrix is indeed a scaling of the one given to us for testing. The estimated location of the camera is approximately the same as well. Both are slightly off, but it is all right since it is not extremely different.
Results for Part 2:


|
The epipolar lines are approximately crossing through the points, showing that the fundamental matrix is a good estimate.
Results for Part 3:
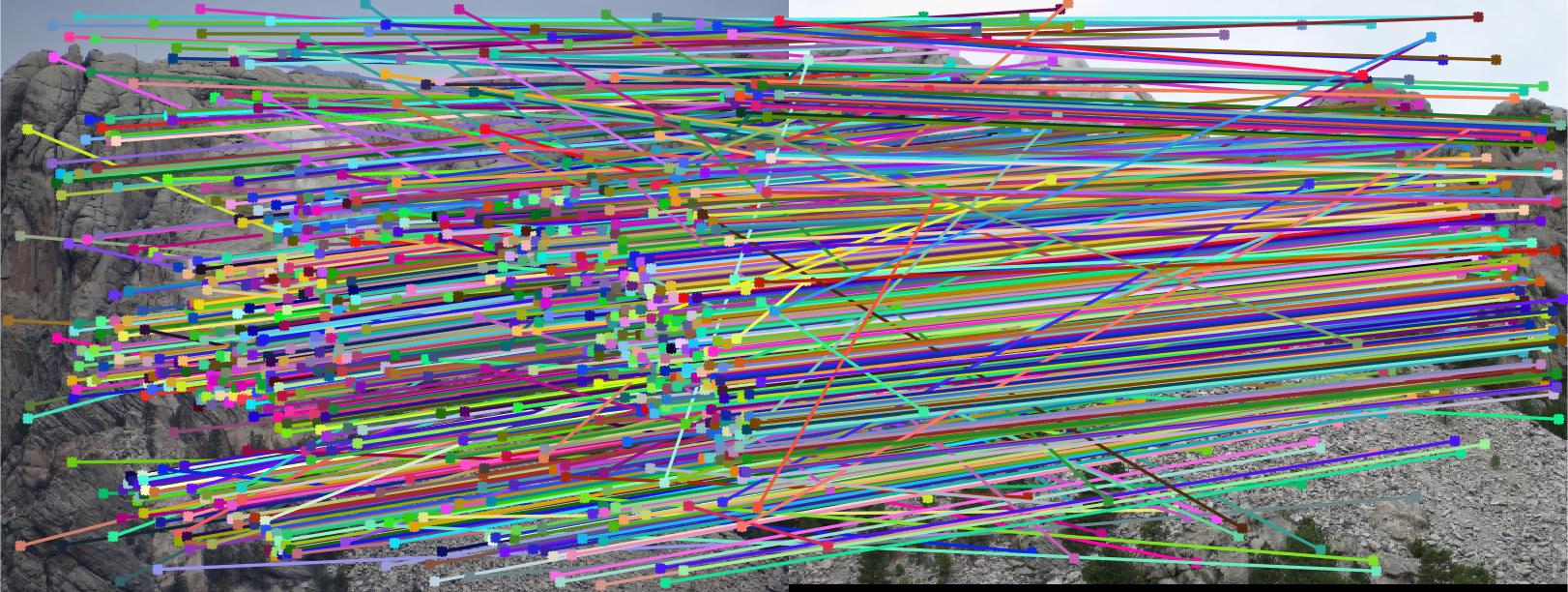
|

|

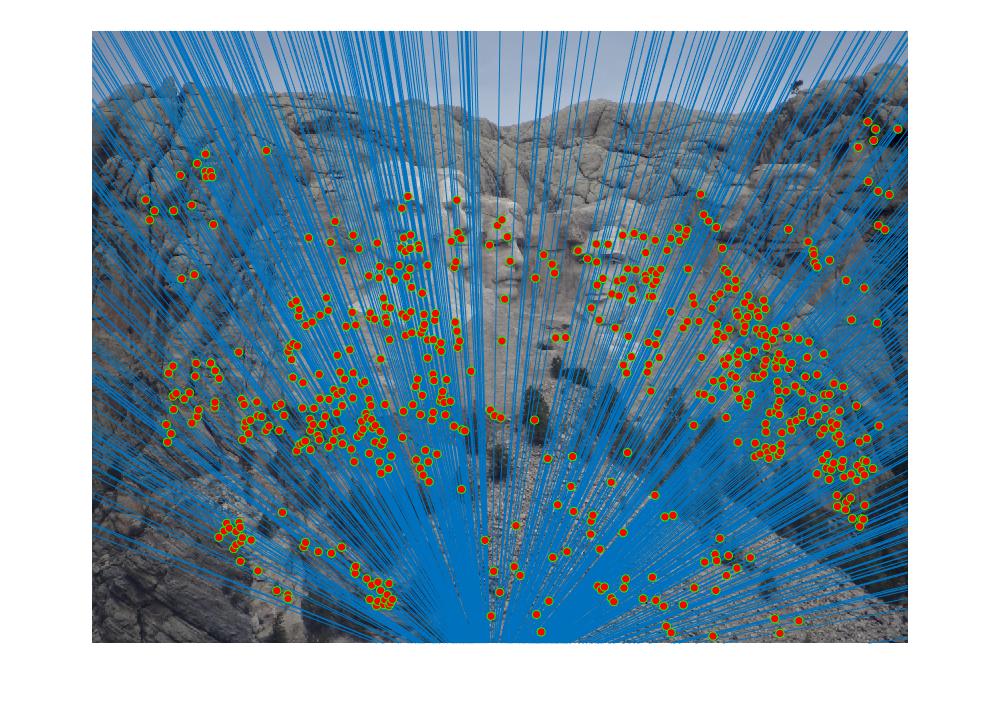
|

|

|
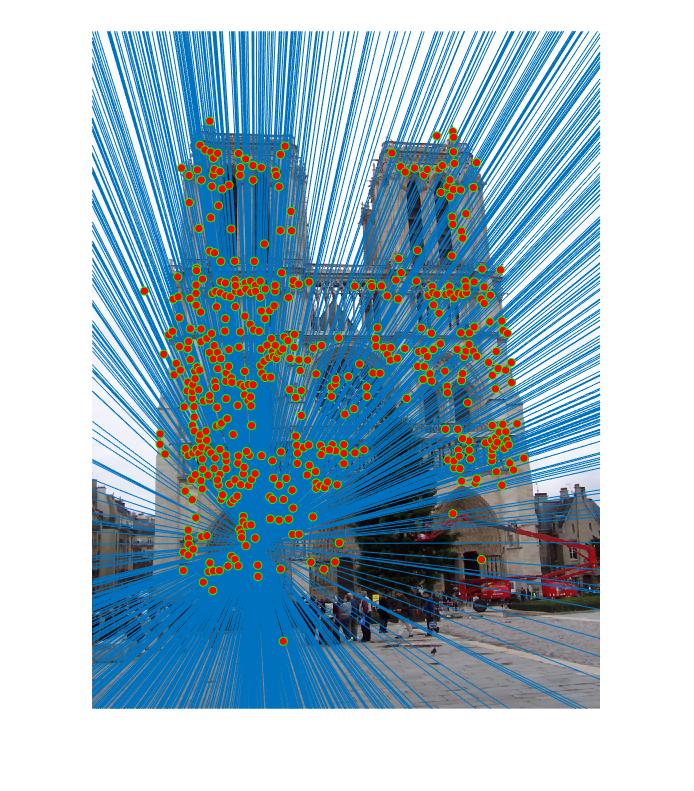

|
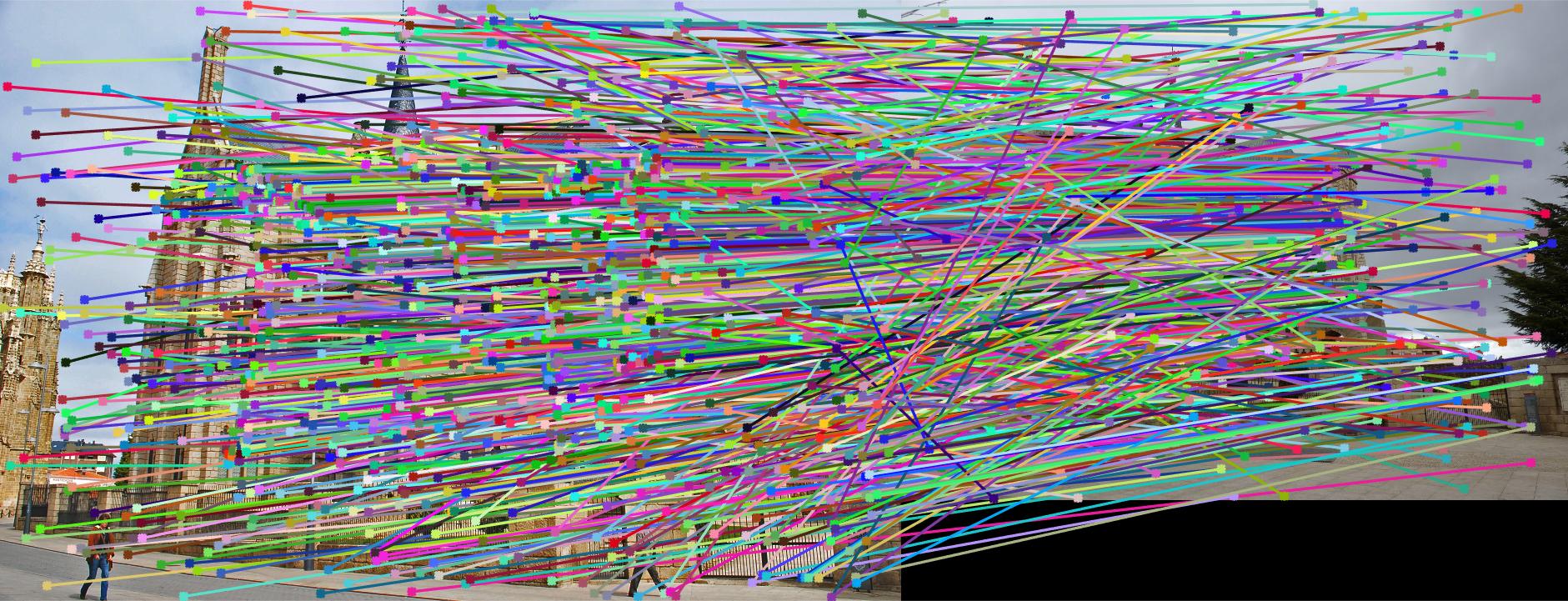
|

|


|

|

|


|
The reason the results may not be good many of the times for some pictures is that I have not normalized the coordinates. This has an effect on Part 2, which also affects Part 3 since RANSAC uses Part 2.