Project 3 / Camera Calibration and Fundamental Matrix Estimation with RANSAC
Visualization of arrows of matching Mount Rushmore.
Objective
The goal of this assignment is to understand camera and scene geometry. In detials, we will learn to estimate the camera projection matrix and fundamental matrix.
The assignment contains several parts:
- Camera Projection Matrix.
- Fundamental Matrix Estimation.
- Fundamental Matrix with RANSAC.
Method and Algorithm
1. Camera Projection Matrix
In this assignment, we are taught to compute the projection matrix that goes from 3D coordinates to 2D's. After rewriting the equation, we get m21X+m22X+m23Z+m24-m31vX-m32vY-m33vZ-m34v=0. And in matrix expression:
[ X1 Y1 Z1 1 0 0 0 0 -u1*X1 -u1*Y1 -u1*Z1; 0 0 0 0 X1 Y1 Z1 1 -v1*Z1 -v1*Y1 -v1*Z1; ... Xn Yn Zn 1 0 0 0 0 -un*Xn -un*Yn -un*Zn; 0 0 0 0 Xn Yn Zn 1 -vn*Zn -vn*Yn -vn*Zn] *[M11; M12; M13; M14; M21; M22; M23; M24; M31; M32; M33]=[u1; v1; u2; v2; ... un vn]
Here I assume M34=1 at first and afer calculating the projection matrix, I normalize it. The result is
[0.0069 -0.0040 -0.0013 -0.8266
0.0015 0.0010 -0.0073 -0.5627
0.0000 0.0000 -0.0000 -0.0034]
2. Fundamental Matrix Estimation
In this assignment, it is required to estimate the mapping of points in one image to lines in another one by means of the fundamental matrix. Here I implement the graduate credit part. Since the input coordinates are 2D, first add one column of 1 to make [u, v, 1] and then normalise it. The goal is to set the centroid of vectors to origin and the mean distance between them and origin as sqrt(2). The transformation matrix will be saved for denormalising use. According to the equation:
[u' v' 1]*[f11 f12 f13; f21 f22 f23; f31 f32 f33]=[u; v; 1]
Compute the constraint matrix A and do singular value decomposition of it. [U,S,V]=svd(A) produces a diagonal matrix S with diagonal elements in descreasing order. So that U and V are unique and V(:,9) is the column with smallest S diagonal element. Construct F using V(:,9) and do another SVD on F to enforce the smallest diagonal element of D to 0. Reconstruct it and denormlise it to fit the provided model using calculated T matrix. The result is:
[ -0.0000 0.0000 -0.0005
0.0000 -0.0000 0.0037
-0.0000 -0.0051 0.1192]
During the test of calculating x'*F*x, I found that that the result ranges from -0.005 to 0.005. So that I set the threshold in part 3 as the 0.005.
3. Fundamental Matrix with RANSAC
In this assignment, it is required to use RANSAC method to find the best estimated fundamental matrix. I randomly choose 8 pairs and calculate the F-matrix using function of part two. Calculate x'*F*x to find inlier points using the threshold based on the test result in part two. After doing such loop 1000 times and find the best estimated fundamental matrix, which has the most inlier points
Explanation of Code
The javascript in thehighlighting folder is configured to do syntax highlighting in code blocks such as the one below.
%Part 1
[N,~]=size(Points_3D);
A = zeros(2*N,11);
B = zeros(2*N,1);
M = zeros(3,4);
for i = 1:N
A(1+2*(i-1),:)=[Points_3D(i,1) Points_3D(i,2) Points_3D(i,3) 1 ...
0 0 0 0 ...
-Points_3D(i,1)*Points_2D(i,1) -Points_3D(i,2)*Points_2D(i,1) ...
-Points_3D(i,3)*Points_2D(i,1)];
B(1+2*(i-1),1)=Points_2D(i,1);
A(2*i,:)=[0 0 0 0 ...
Points_3D(i,1) Points_3D(i,2) Points_3D(i,3) 1 ...
-Points_3D(i,1)*Points_2D(i,2) -Points_3D(i,2)*Points_2D(i,2) ...
-Points_3D(i,3)*Points_2D(i,2)];
B(2*i,1)=Points_2D(i,2);
end
M_tmp = A\B;
M_tmp = [M_tmp;1];
for i = 1:3
M(i,:) = M_tmp((4*(i-1)+1):(4*(i-1)+4));
end
M = -M/sqrt((sum(sum(M.^2))));
%Part 2
%%%%Normalise
[M,N]=size(point);
tmp = zeros(M,N);
%Set centroid
centroid = mean(point(:,1:2));
tmp(:,1) = point(:,1)-centroid(1);
tmp(:,2) = point(:,2)-centroid(2);
%calculate distance
dist = sqrt(tmp(:,1).^2 + tmp(:,2).^2);
meandist = mean(dist);
%set mean distance to sqrt2
scale = sqrt(2)/meandist;
T = [scale 0 -scale*centroid(1)
0 scale -scale*centroid(2)
0 0 1 ];
point_norm = (T*point')';
%%%%Estimate fundamental matrix%%%%
[N,~]=size(Points_a);
A = zeros(N,9);
P_a = [Points_a ones(N,1)];
P_b = [Points_b ones(N,1)];
[P_a,T1] = normalise(P_a);
[P_b,T2] = normalise(P_b);
P_a = P_a(:,1:2);
P_b = P_b(:,1:2);
for i = 1:N
A(i,:)=[P_a(i,1)*P_b(i,1) P_a(i,2)*P_b(i,1) ...
P_b(i,1) P_a(i,1)*P_b(i,2) ...
P_a(i,2)*P_b(i,2) P_b(i,2) P_a(i,1) ...
P_a(i,2) 1];
end
[U,S,V]=svd(A,0);
F = reshape(V(:,9),3,3)';
[FU,FS,FV]=svd(F,0);
FS(3,3)=0;
F=FU*FS*FV';
F = T2'*F*T1;
% F=P_b_H*F*P_a_H';
F_matrix = F;
%Part 3
[M,N] = size(matches_a);
inlierNum = 0;
for I = 1:1000
random = randsample(M,8);
M_a = matches_a(random,:);
M_b = matches_b(random,:);
Fmatrix = estimate_fundamental_matrix(M_a,M_b);
inlier = [];
for i = 1:M
if abs([matches_b(i,:) 1]*Fmatrix*[matches_a(i,:) 1]')<0.005
inlier = [inlier i];
else
end
end
if length(inlier)>inlierNum
inlierBest = inlier;
inlierNum = length(inlier);
Best_Fmatrix = Fmatrix;
else
end
end
inliers_a = matches_a(inlierBest,:);
inliers_b = matches_b(inlierBest,:);
Results in a table
Part2 result

|

|
Part3 Notre Dame

|

|
Part3 Mount Rushmore
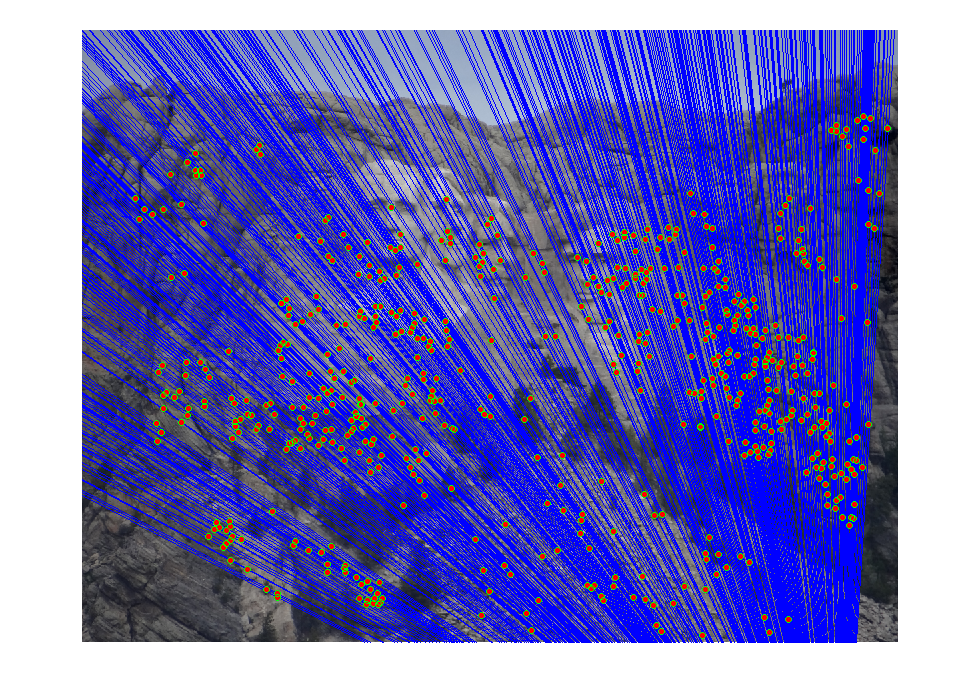
|
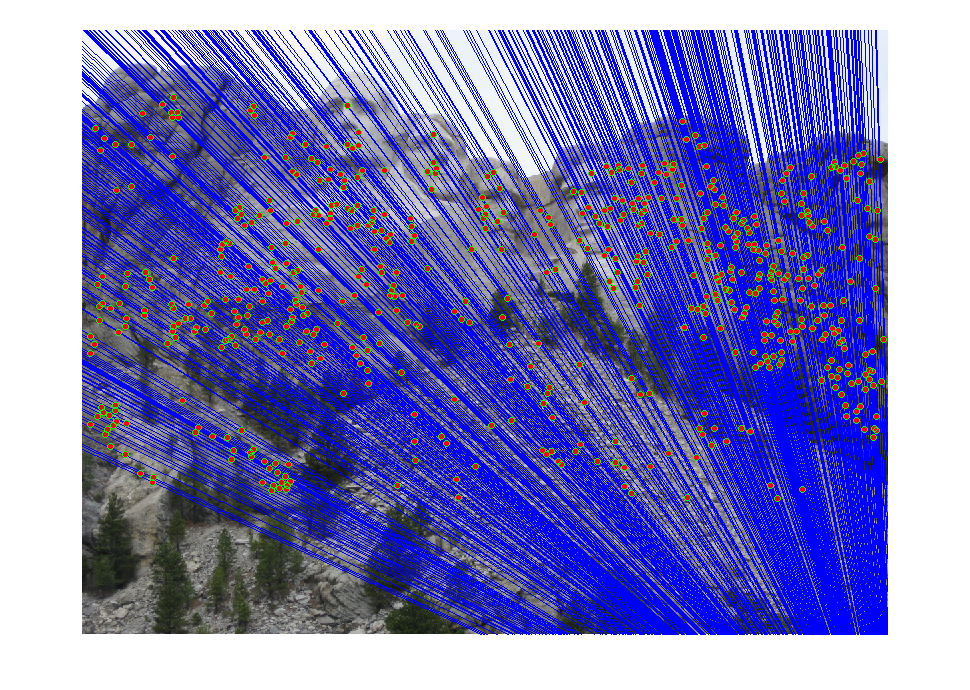
|
Part3 Episcopal Gaudi
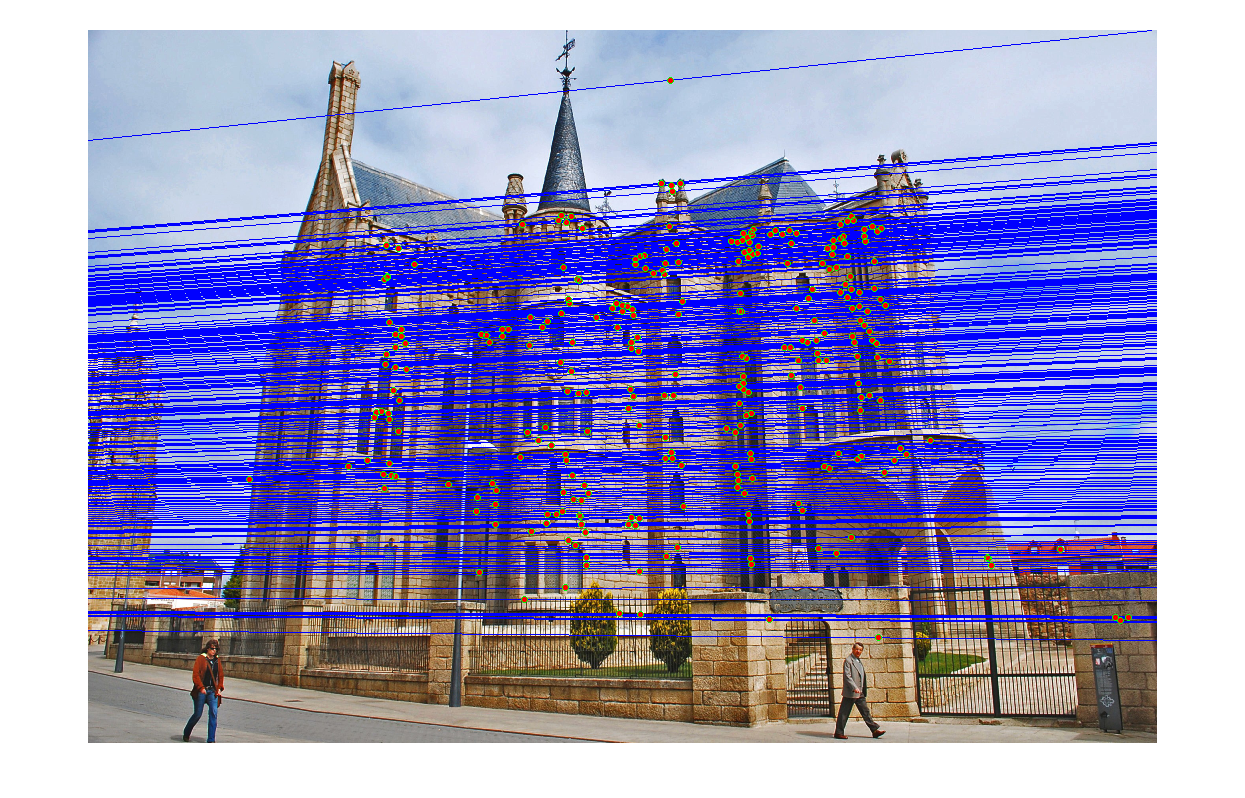
|

|