I. Camera Projection matrix estimation
This function determines the projection matrix, which is a 3 X 4 matrix that describes the mapping of a pinhole camera from 3D points in the world to 2D points in an image. A homogeneous linear system of equations for moving from 3D world coordinates to 2D image coordinates using the projection matrix, M is considered. A set of corresponding normalized 2D and 3D points are substituted in the equations and the unknown values of M are determined. The total residual between the projected 2D location of each 3D point and the actual location of that point in 2D image must be very small. This projection matrix M can be divided into a matrix K of intrinsic parameters and a matrix [R/T] of extrinsic parameters. Camera centre is then determined as -(Q^-1)*m4; where m4 is the last column of M and Q is the first 3X3 elements of M.
Sample Output
The projection matrix is :
| 0.7679 | -0.4938 | -0.0234 | 0.0067 |
| -0.0852 | -0.0915 | -0.9065 | -0.0878 |
| 0.1827 | 0.2988 | -0.0742 | 1.0000 |
The total residual is: 0.0445.
The estimated location of camera is: -1.5126, -2.3517, 0.2827
II. Fundamental matrix estimation
To obtain the epipolar geometry, we need to estimate fundamental matrix F. This can be done using some initial point correspondences between the images. Equation below shows that each matching pair of points between the two images provides a single linear constraint on F. This allows F to be estimated linearly (up to the usual arbitrary scale factor) from 8 independent correspondences.
(f11uu’ + f12vu’ + f13u’ + f21uv’ + f22vv’ + f23v’ + f31u + f32v + f33) = 0, where u,v,u’ and v’ are corresponding point locations in two images.
The linear squares estimate of F is full rank; however, the fundamental matrix is a rank 2 matrix. In order to reduce the rank, we can decompose F using singular value decomposition into the matrices U ΣV' = F. We can then estimate a rank 2 matrix by setting the smallest singular value in Σ to zero thus generating Σ2 . The fundamental matrix is then easily calculated as F = U Σ2 V'.
Output


|
III. Fundamental matrix with RANSAC
The fundamental matrix is computed with unreliable point correspondences computed with SIFT. Least squares regression is not appropriate in this scenario due to the presence of multiple outliers. RANSAC is used in order to determine the fundamental matrix. 8 random corresponding matched points from two images are taken and the fundamental matrix is solved and determined. The number of inliers for this fundamental matrix is computed. To determine the points that correspond to inliers for a particular fundamental matrix, a distance measure or threshold is taken. Thus, a threshold is picked between the inliers and outliers and this threshold is varied in order to get better results.This process is iteratively done and the fundamental matrix with the highest number of inliers is taken as the final fundamental matrix.
Example code
Output

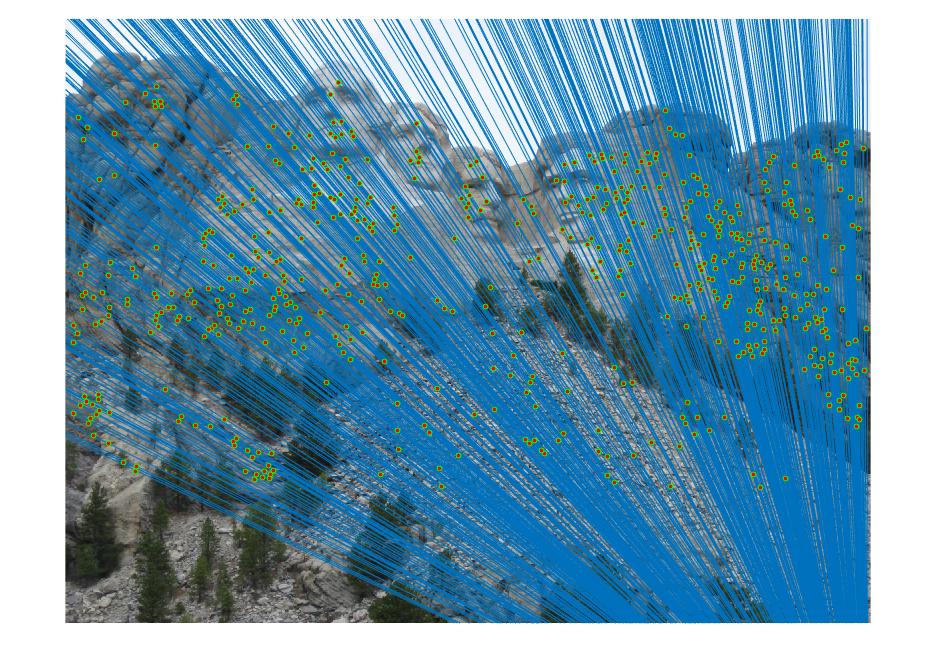
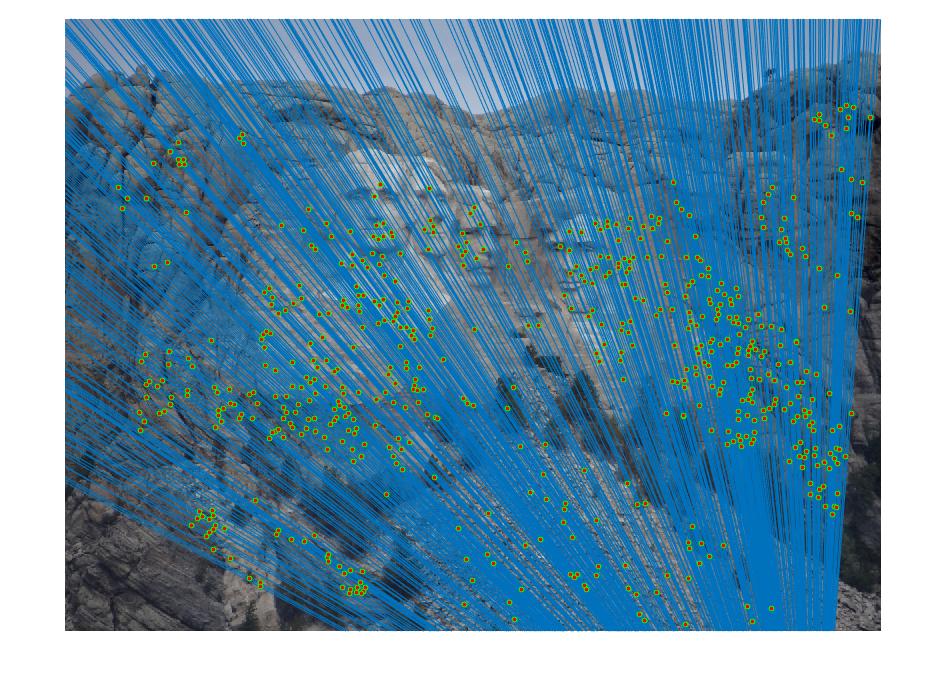
|
IV. Extra Credit: Fundamental matrix estimation after normalizing coordinates
Estimate of the fundamental matrix can be improved by normalizing the coordinates before computing the fundamental matrix. Normalization is performed through linear transformations to make the mean of the points zero and the average magnitude about 1.0 or some other small number.
Example code
c_u_a = mean(Points_a(:,1)); %finding the mean of points
c_v_a = mean(Points_a(:,2));
c_u_b = mean(Points_b(:,1));
c_v_b = mean(Points_b(:,2));
offset_a = eye(3) + [zeros(3,2) [-c_u_a ; -c_v_a ; 1]];
offset_b = eye(3) + [zeros(3,2) [-c_u_b ; -c_v_b ; 1]];
s_u_a = 1/std(Points_a(:,1));
s_v_a = 1/std(Points_a(:,2));
s_u_b = 1/std(Points_b(:,1));
s_v_b = 1/std(Points_b(:,2));
scale_a = [s_u_a , 0 , 0 ; 0 , s_v_a , 0 ; 0 ,0 , 1]; %scale matric is determined by using inverse of standard deviation
scale_b = [s_u_b , 0 , 0 ; 0 , s_v_b , 0 ; 0 ,0 , 1];
transform_a = scale_a * offset_a; %transform matrix is the product of scale and offset matrices
transform_b = scale_b * offset_b;
[n m]=size(Points_a);
output_a = transform_a * [ Points_a ones(n,1) ]';
output_b = transform_b * [ Points_b ones(n,1) ]';
output_a = output_a';
output_b = output_b';
w=zeros(n,9);
w = [output_a(:,1).*output_b(:,1) output_a(:,2).*output_b(:,1) output_b(:,1) output_a(:,1).*output_b(:,2) output_a(:,2).*output_b(:,2) output_b(:,2) output_a(:,1) output_a(:,2) ones(n,1)];
[U, S, V] = svd(w);
q = V(:,9);
q=reshape(q',3,3);
[u, s, v] = svd(q);
s(3,3) = 0;
F_matrixnorm = u*s*v';
F_matrix = transform_b' * F_matrixnorm * transform_a;
Output
Notre Dame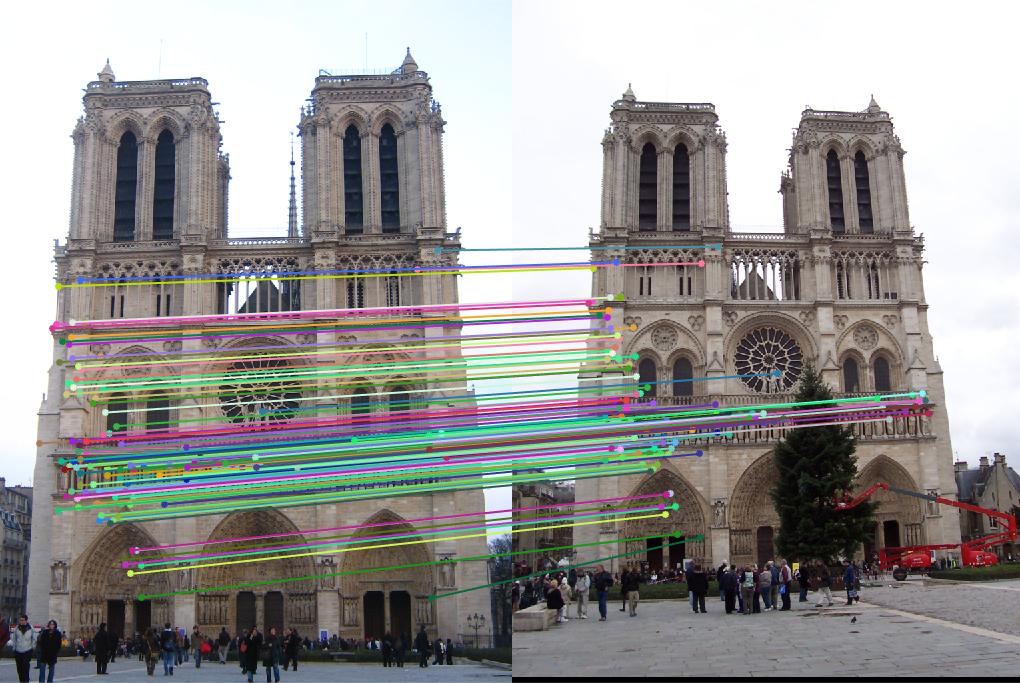


|
Episcopal Gaudi


|
Mount Rushmore with threshold 0.05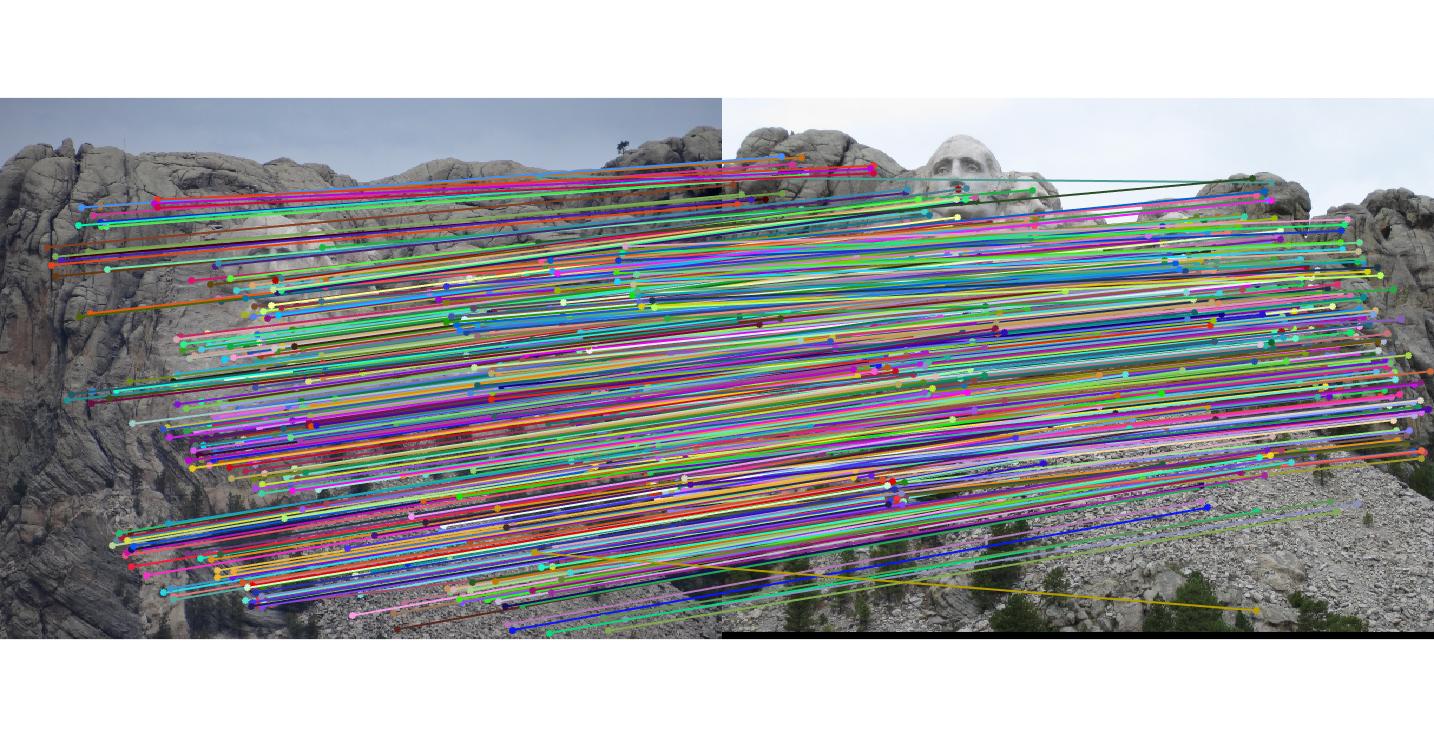
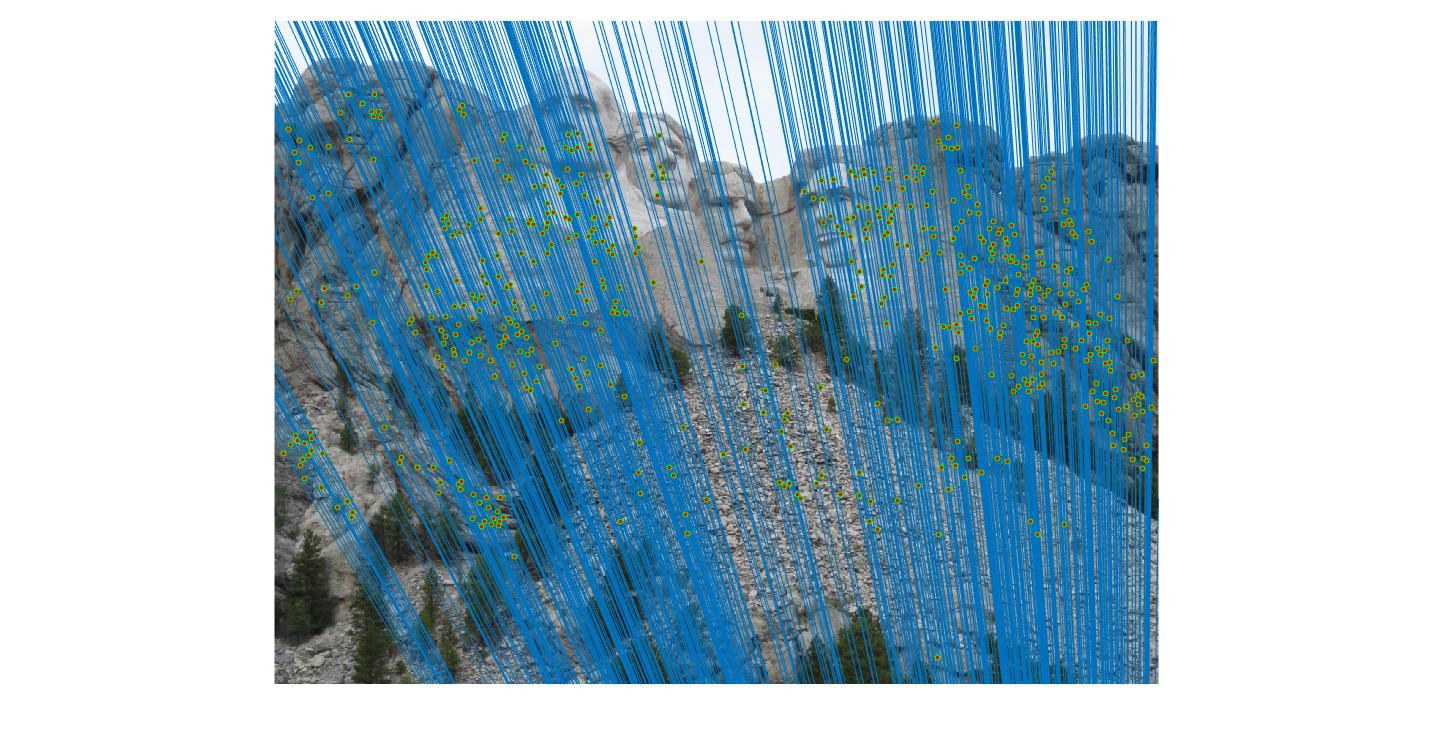
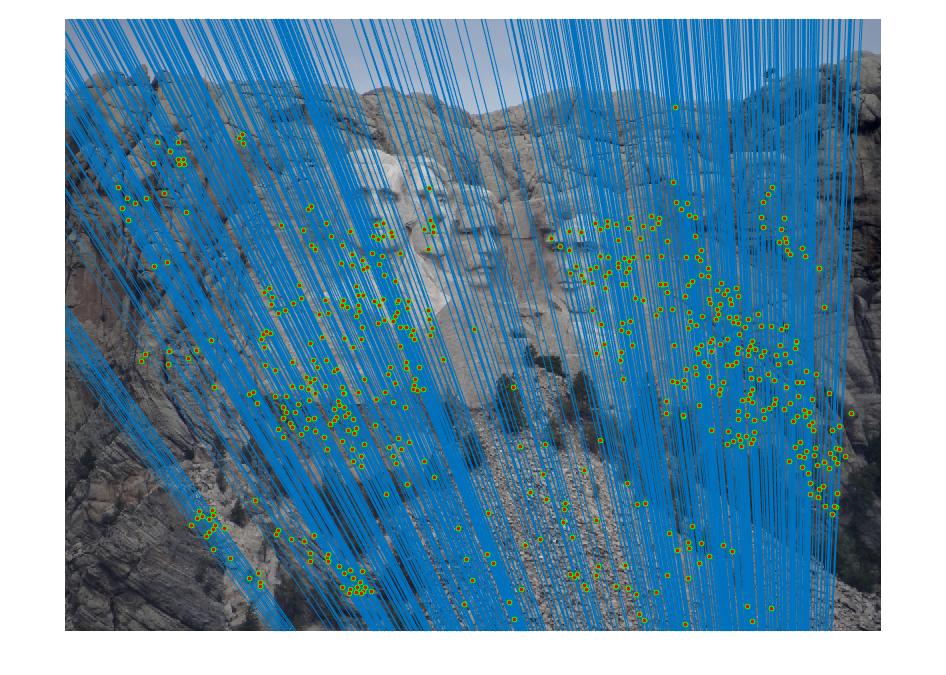
|
Mount Rushmore with threshold 0.005


|
By using RANSAC to find the fundamental matrix with the most inliers, spurious matches can be filtered away and near perfect point to point matching can be acheived.