Project 3: Camera Calibration and Fundamental Matrix Estimation with RANSAC
The objective of this project is to estimate the camera projection matrix and the fundamental matrix using RANSAC.
Part 1
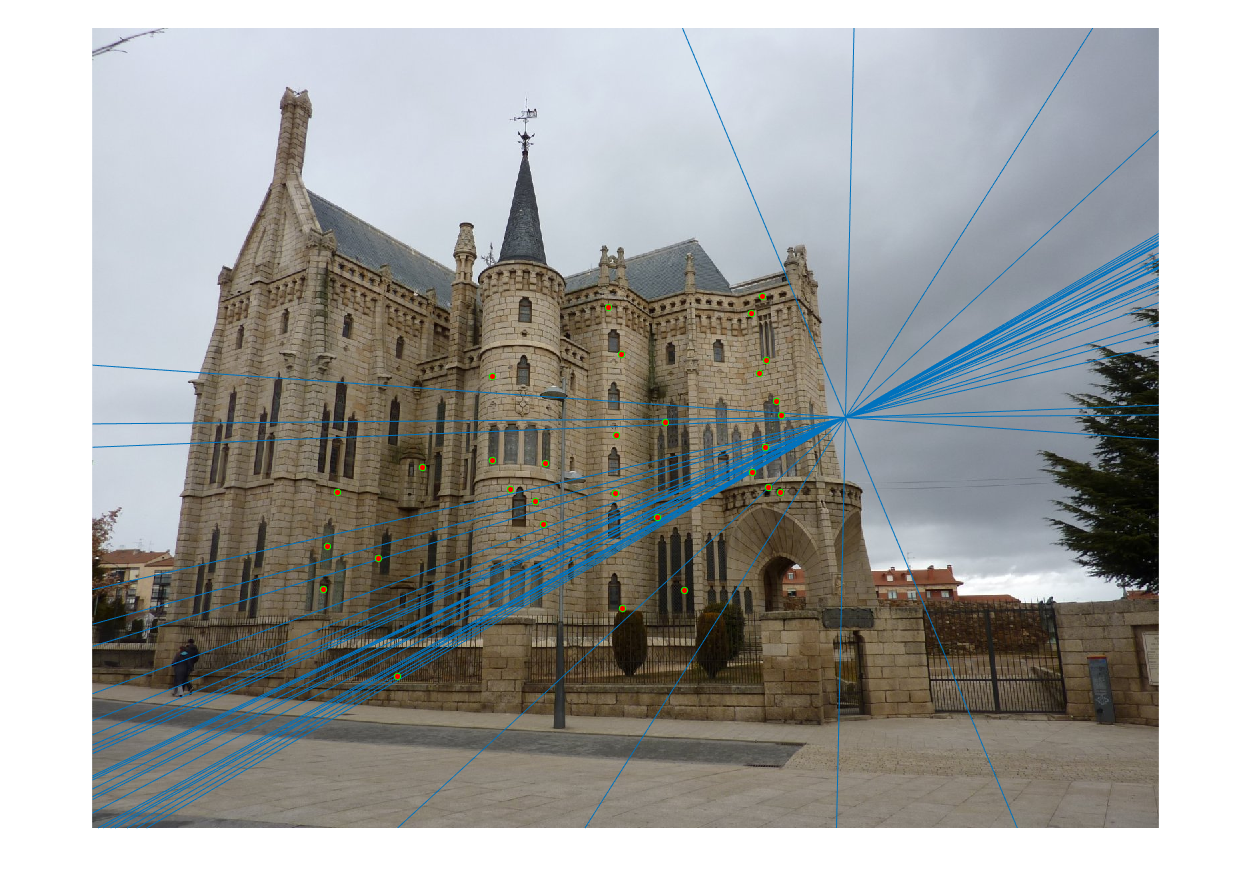
The first task involved in this project involves finding the camera projection matrix that maps from 3D world coordinates to 2D images coordinates. A major component of this is finding the M matrix. In order to fix a scale for the M matrix, we can either set the last element of M (m34) equal to 1 or use Single Value Decomposition and solve the optimization function. The matrix M is solved for by using least squares regression
This is simple to implement in MATLAB by solving a set of linear equations. In order to fix the scale and obtain the unique solution, I use Single Variable Decomposition through a svd() call. My total residual for my implementation is 0.0445.
function M = calculate_projection_matrix( Points_2D, Points_3D )
[n, ~] = size(Points_2D);
A = zeros(2*n, 12);
for i = 1: n
d2 = Points_2D(i, :);
d3 = Points_3D(i, :);
u = d2(1, 1);
v = d2(1, 2);
rows = [d3 1 0 0 0 0 -u.*d3 -u; 0 0 0 0 d3 1 -v.*d3 -v];
A(2*i-1:2*i, :) = rows;
end
[~, ~, V] = svd(A);
M = V(:,end);
M = reshape(M,[],3)';
end
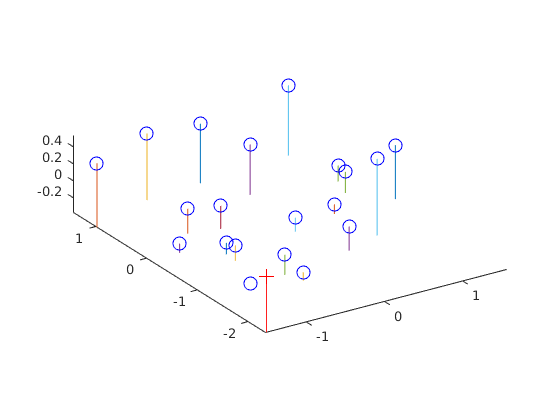
Once we have M, we can estimate the camera's position using the equation C = -Q^(-1)*m4. This is straighforward in MATLAB. The estimated location of the camera is [-1.5127, -2.3517, 0.2826].
function [ Center ] = compute_camera_center( M )
Q = M(1:3,1:3); % C = -Q^(-1)*m4
m4 = M(:,4);
C = -inv(Q)*m4;
Center = C;
end
Part 2
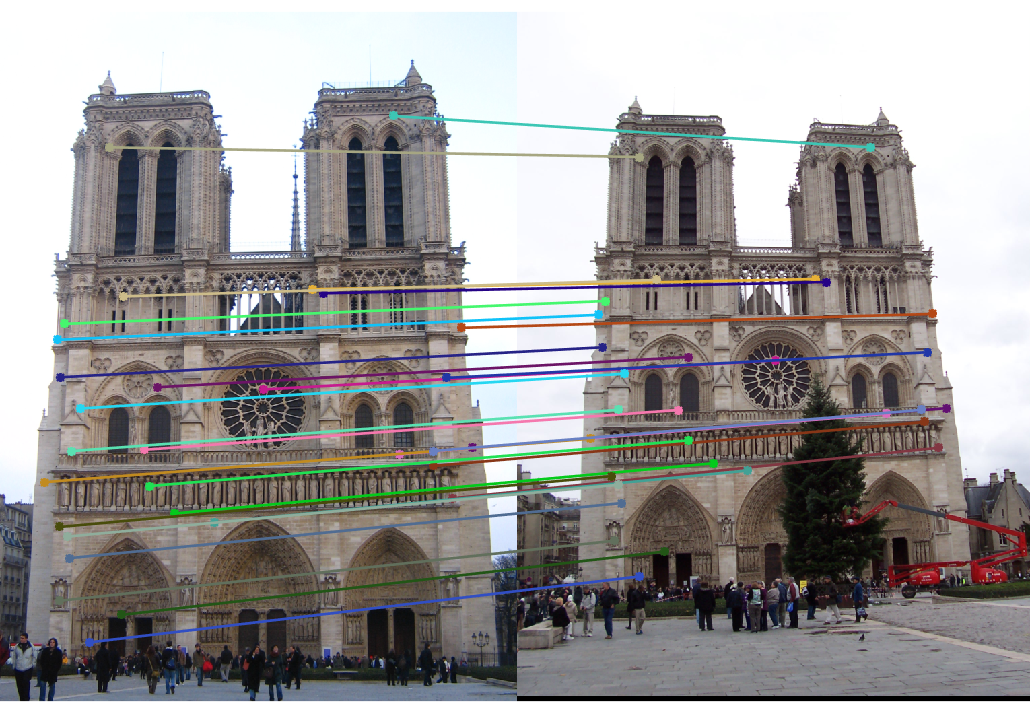
The second task is to estimate the Fundamental Matrix. This fundamental matrix is found using similar methods, like SVD and setting the scale prior to solving the regression, used in Part 1.
The estimated projection matrix using pic_a and pic_b is: [-0, 0, -0.0019; 0, 0, 0.0172; -0.0009, -0.0264, 0.9995]
function [ F_matrix ] = estimate_fundamental_matrix(Points_a,Points_b)
[n, ~] = size(Points_a);
A = zeros(n, 9);
for i = 1: n
tempA = Points_a(i, :);
tempB = Points_b(i, :);
uA = tempB(1, 1);
vA = tempB(1, 2);
uB = tempA(1, 1);
vB = tempA(1, 2);
A(i, :) = [uA*uB uA*vB uA vA*uB vA*vB vA uB vB 1];
end
[~, ~, V] = svd(A);
F = V(:, end);
F_matrix = reshape(F, [3 3])';
[U, S, V] = svd(F_matrix);
S(3, 3) = 0;
F_matrix = U * S * V';
end


|
Part 3
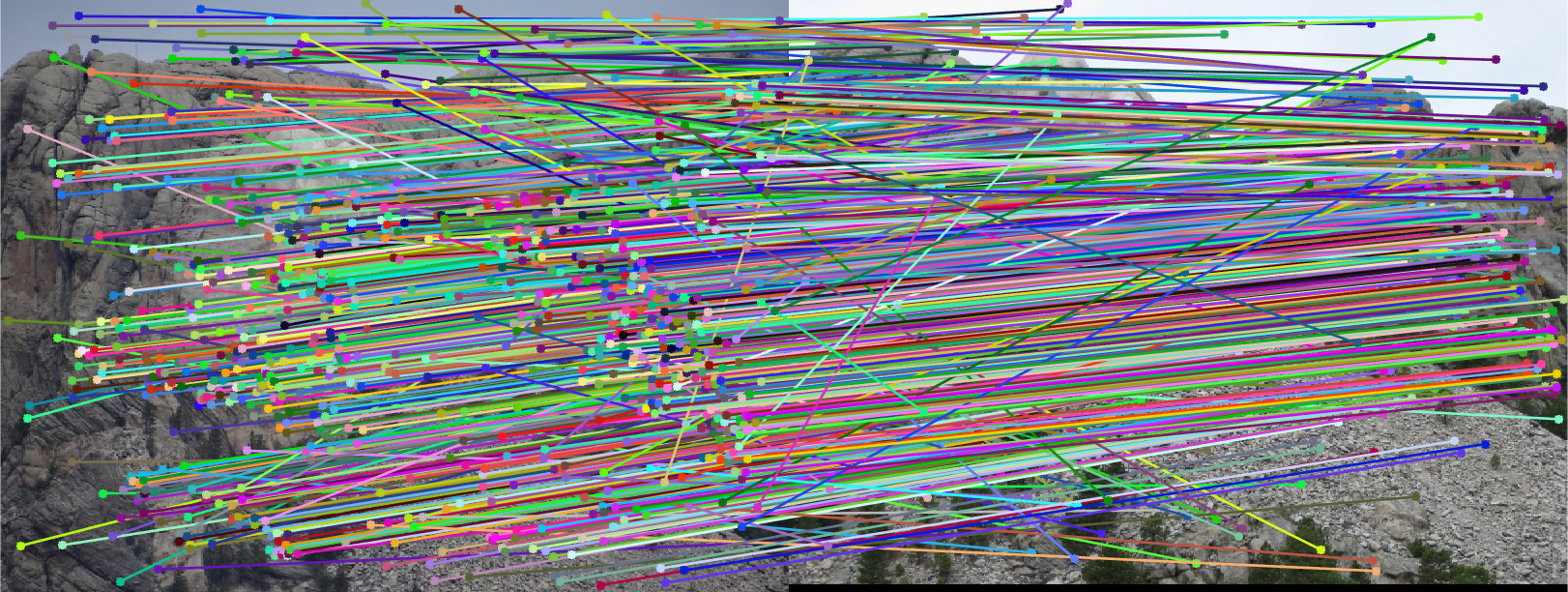
The last task is to make our fundamental matrix more robust to point correspondence with the use of RANSAC. RANSAC is a great tool in the case of there being outliers. We find a random subset of matching pairs through a large number of iterations. With each iteration, points are classified as inliers when the product of their multiplication with the fundamental matrix is small compared with outliers. I found a threshold of 0.3 to work well. I chose to use 10,000 iterations in this implementation.
function [ Best_Fmatrix, inliers_a, inliers_b] = ransac_fundamental_matrix(matches_a, matches_b)
numMatches = size(matches_a,1);
maxCount = 0;
for i = 1:10000
thisSample = randsample(numMatches,8);
Fmatrix = estimate_fundamental_matrix(matches_a(thisSample,:), matches_b(thisSample,:));
matchA = [];
matchB = [];
count = 0;
for n = 1:numMatches
error = [matches_a(n,:) 1]*Fmatrix'*[matches_b(n,:) 1]';
if abs(error) < 0.03
matchA(end+1,:) = matches_a(n,:);
matchB(end+1,:) = matches_b(n,:);
count = count + 1;
end
end
if count > maxCount
maxCount = count;
Best_Fmatrix = Fmatrix;
inliers_a = matchA;
inliers_b = matchB;
end
end
pool = randsample(size(inliers_a,1),30);
inliers_a = inliers_a(pool,:);
inliers_b = inliers_b(pool,:);
end
|

|


|
|


|

|

|