Project 3 Camera Calibration and Fundamental Matrix Estimation with RANSAC
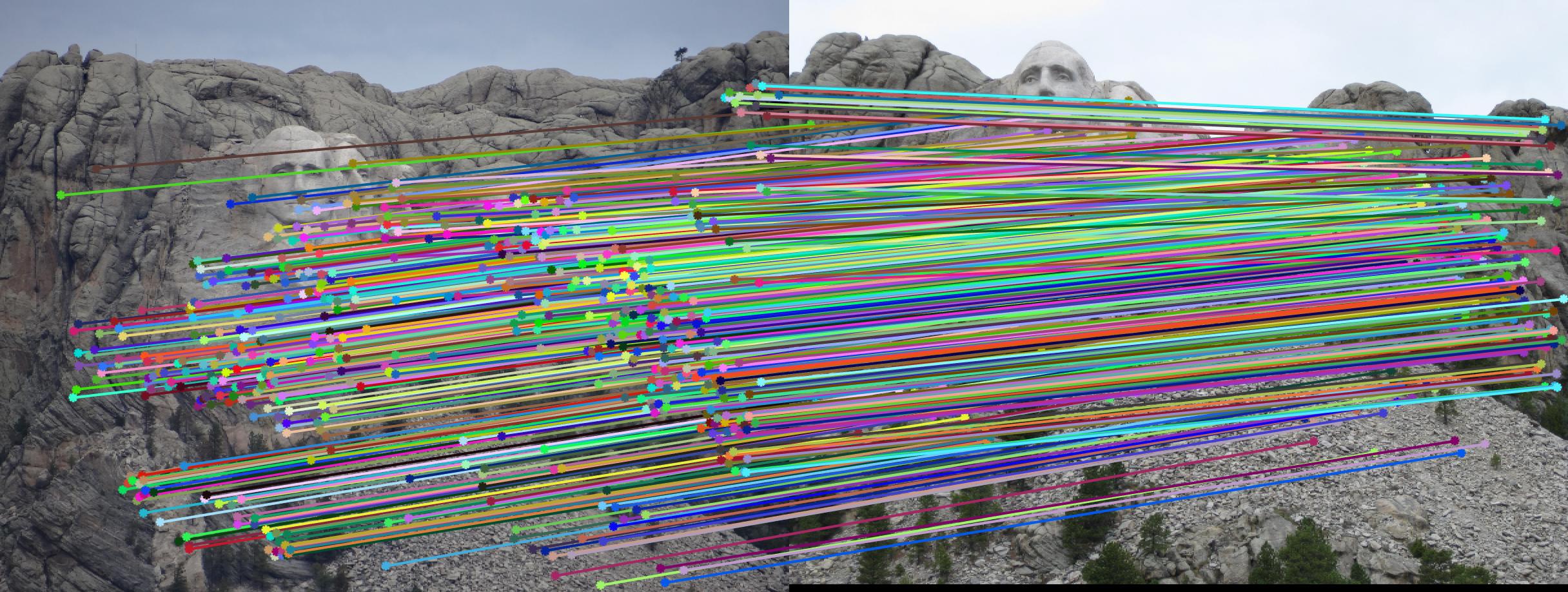
Mount Rushmore match points.
Project Part 1: Estimate the projection matrix and find camera center
The project is divided into 3 parts. The first one is to find camera projection matrix. The specific steps are as follows:
- Two functions need to be implemented in this part: Calculate the projection matrix and find the camera center.
- For projection matrix, set up the homogeneous linear system using the normalized projection points from 3d to 2d.
- Solve the extrinsic transformation matrix by using singular value decompositions.
- Calculate the center of the camera using the equation: C = -Q^-1*m4.
Project Part 2: Fundamental Matrix Estimation
The second part of the project is to estimate the fundamental matrix, which is used to map the points from one image to corresponding points in the other image
- Construct the homogeneous linear system the same way as in part 1.
- Reduce the rank of the fundamental matrix by setting the last column of the singular value of Σ.
- Reevaluate the fundamental matrix as described on the website.
Initially when doing the normalized patch as feature descriptions, the accuracy jumped from nearly 0% to 42%. When SIFT-life feature is implemented, the accuracy rose up to 66% for Notre Dame pair.
Project Part 3: Fundamental Matrix with RANSAC
In this part, we are going to combine the fundamental matrix evaluation function with RANSAC algorithm
- Use SIFT matching to find potential matching points between two images
- For each iteration, randomly select 8 match pairs to construct the fundamental matrix.
- search for all the pairs of the potential matches and evaluate whether each pair is within the RANSAC threshold.
- Count the number of points fall in the range, if the number is larger than threshold,jump out of the loop.
- Record the fundamental matrix that capture most of the matching points.
Code for match_features.m
for i = 1:iterations
sample = randsample(row,8);
samples_matches_a = matches_a(sample,:);
samples_matches_b = matches_b(sample,:);
temp_F_matrix = estimate_fundamental_matrix(samples_matches_a,samples_matches_b);
temp_inliers_a = [];
temp_inliers_b = [];
num_matches = 0;
for j=1:row
if abs([matches_b(j, :) 1] * temp_F_matrix * [matches_a(j, :) 1]') < inlier_distance_threshold
temp_inliers_a = [temp_inliers_a;matches_a(j,:)];
temp_inliers_b = [temp_inliers_b;matches_b(j,:)];
num_matches = num_matches + 1;
end
end
if (num_matches / row) >= threshold
max_a = temp_inliers_a;
max_b = temp_inliers_b;
break
elseif max_inlines < num_matches
max_inlines = num_matches;
max_a = temp_inliers_a;
max_b = temp_inliers_b;
end
end
inliers_a = max_a;
inliers_b = max_b;
Best_Fmatrix = estimate_fundamental_matrix(inliers_a,inliers_b);
Results in a table



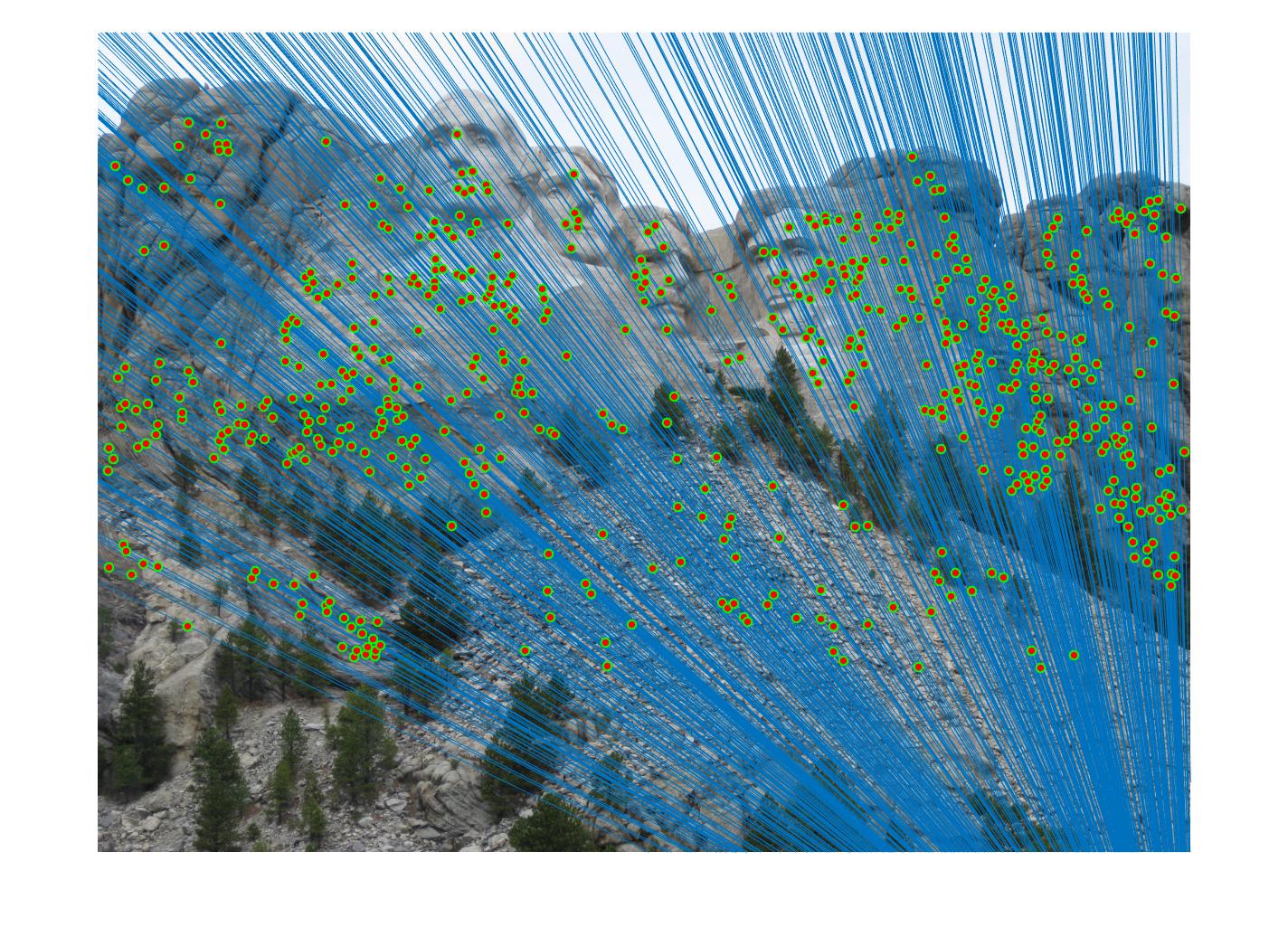
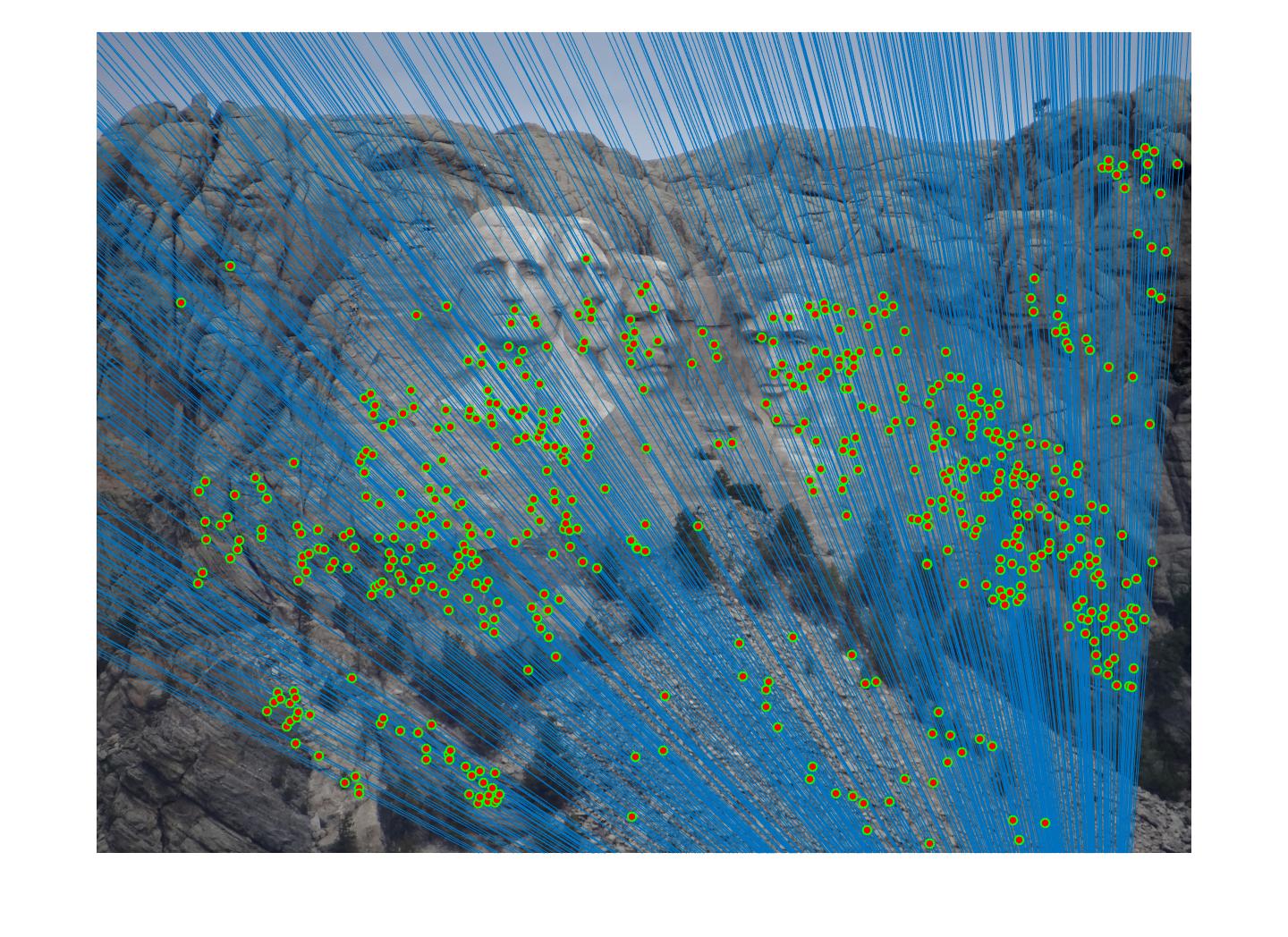
|
For RANSAC algorithm, I found it's very hard to get over certain thresholds, say 60 or 70 percent. Therefore, the code returns the matrix that returns most agreeable points. When threshold gets too big,for example, above 0.1, there will be obvious mismatch pairs get selected.