Project 3 / Camera Calibration and Fundamental Matrix Estimation with RANSAC
In this project, we are going to complete three tasks. First, we will estimate the camera projection matrix as well as calculate the camera center, given a group of mapping from 3D world coordinates to 2D image coordinates. Then, we will estimate the fundamental matrix which relates epipolar lines in two images. Finally, we will use RANSAC to find the 'best' fundamental matrix with the most inliers.
Algorithm
Part1
From given points correspondence, we can use following equations to calculate the projection matrix M.
M_11X + M_12Y + M_13Z + M_14 - M_31uX -M_32uY - M_33uZ - M_34u = 0
M_21X + M_22Y + M_23Z + M_24 - M_31vX -M_32vY - M_33vZ - M_34v = 0
Then we can use SVD to solve this, and the camera center will be C = -Q^(-1) * m_4.
Part2
The fundamental marix could be calculated in similar way as in Part 1. From given 2D points, we have following equation
u'uf_11 + u'vf_12 + u'f_13 + v'uf_21 + v'vf_22 + v'f_23 + uf_31 + vf_32 + f_33 = 0
Use SVD, we get the fundamental matrix F. Since F is full rank, but the fundamental should be a rank 2 matrix. So I decomposed F and set the smallest singular value to be zero.
Part3
In this part, I randomly chose 9 pairs of points and use the function in Part 2 to estimate the fundamental matrix. Then compare the number of inliers with the current best solution. If it's better, update it as the best solution. I set the maximum iteration rounds to 10000.
Extra credit
For extra credit, I normalized the coordinates before computing the fundamental matrix. I subtracted the mean of given points from these points, then use the reciprocal as the scale factor. After I calculated the fundamental matrix F, I used F = transpose(T_b) * F * T_a to adjust it so that the F can operate on the original coordinates.
Implementation
Here is my code.
% Part 1
% construct linear regression
for row = 1 : 2 : 2 * point_num
point = ceil(row / 2);
A(row, :) = [Points_3D(point, :), 1, 0, 0, 0, 0, (-1 * Points_2D(point, 1)) * Points_3D(point, :), -1 * Points_2D(point, 1)];
A(row+1, :) = [0, 0, 0, 0, Points_3D(point, :), 1, (-1 * Points_2D(point, 2)) * Points_3D(point, :),-1 * Points_2D(point, 2)];
end
% recover the camera center
Center = -1 * inv(Q) * m4;
% Part 2
% construct linear regression
for i = 1 : num_a
A(i, :) = [Points_a(i, 1) * Points_b(i, 1), Points_b(i, 1) * Points_a(i, 2), Points_b(i, 1),
Points_b(i, 2) * Points_a(i, 1), Points_a(i, 2) * Points_b(i, 2), Points_b(i, 2),
Points_a(i, 1), Points_a(i, 2), 1];
end
% decompose F matrix and set the smallest singular value to zero
[U, S, V] = svd(F_matrix);
[~, ind] = min(S(:));
S2(ind2sub(size(S), ind)) = 0;
F_matrix = U * S * V';
% adjust the fundamental matrix
F_matrix = transpose(S_b * T_b) * F_matrix * (S_a * T_a);
% Part 3
for iteration = 1 : max_iter
% get 9 sample points
idx = randperm(num_points, 9);
points_a = matches_a(idx, :);
points_b = matches_b(idx, :);
% calculate the F matrix
F = estimate_fundamental_matrix(points_a, points_b);
% count inliers
temp_a = [];
temp_b = [];
num_inlier = 0;
for i = 1 : num_points
if (abs([matches_b(i, :), 1] * F * transpose([matches_a(i, :), 1])) <= threshold)
num_inlier = num_inlier + 1;
temp_a = vertcat(temp_a, matches_a(i, :));
temp_b = vertcat(temp_b, matches_b(i, :));
end
end
% update best case
if(num_inlier > max_inlier)
max_inlier = num_inlier;
inliers_a = temp_a;
inliers_b = temp_b;
Best_Fmatrix = F;
end
end
% Extra credit
% normalize points
num_a = size(Points_a, 1);
m_a = mean(Points_a);
T_a = [1, 0, -1 * m_a(1); 0, 1, -1 * m_a(2); 0, 0, 1];
temp = Points_a;
temp = temp - repmat(m_a, num_a, 1);
s = 1 / std2(temp);
S_a = [s, 0, 0; 0, s, 0; 0, 0, 1];
temp = zeros(num_a, 3);
for i = 1 : num_a
temp(i, :) = S_a * T_a * [Points_a(i, 1); Points_a(i, 2); 1];
end
Points_a = temp(:, 1 : 2);
Results
Part1
The projection matrix I calculated was:
[0.4583 -0.2947 -0.0140 0.0040 -0.0509 -0.0546 -0.5411 -0.0524 0.1090 0.1783 -0.0443 0.5968]
The total residual was 0.0445, which was small. The recovered camera center was <-1.5127, -2.3517, 0.2826>. It was not exactly as the same as given expected data, but it was very close. The following pictures show us the acutal points match the projection points.
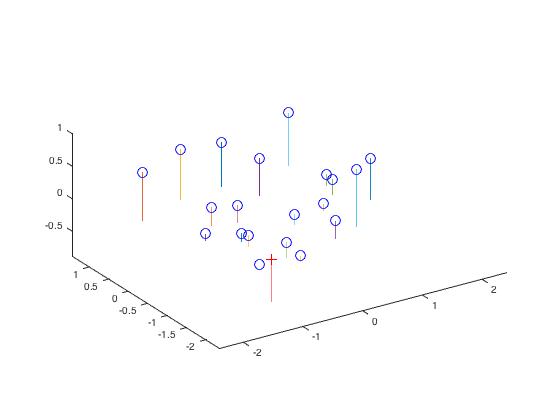
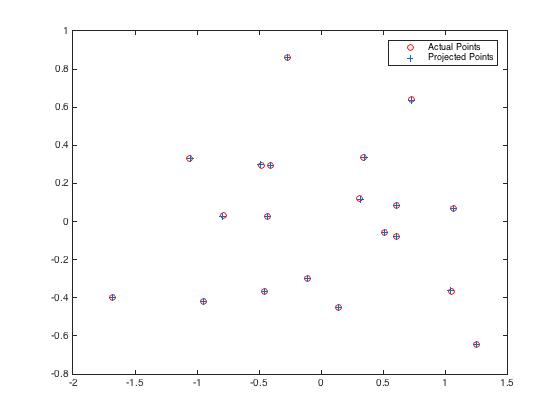
|
Part2
The fundamental matrix was:
[-0.0000 0.0000 -0.0019 0.0000 0.0000 0.0172 -0.0009 -0.0264 0.9995]
When I normalized the points, the fundamental matrix was:
[-0.0000 0.0000 -0.0004 0.0000 -0.0000 0.0032 -0.0000 -0.0044 0.1063]
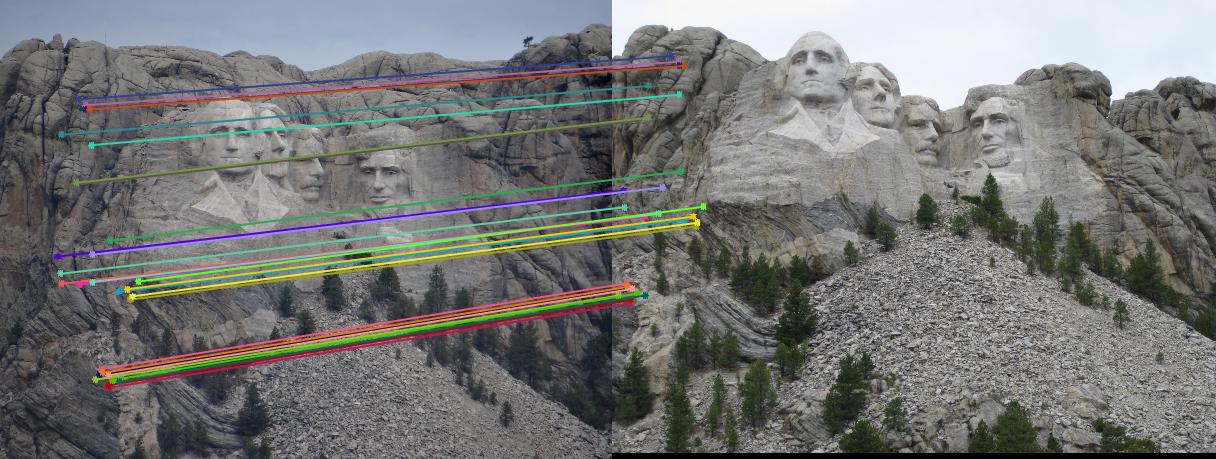
From two pictures shown below, we can see that all matching points were on the corresponding epipolar lines in both scene. There was not much difference before and after normalization.


|
Part 3
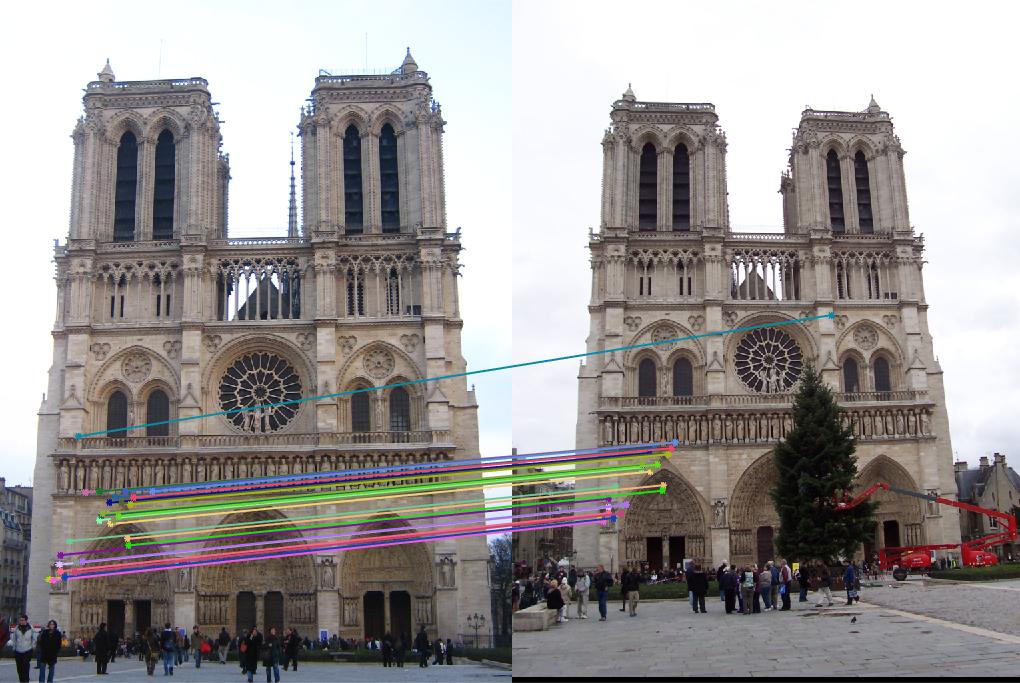
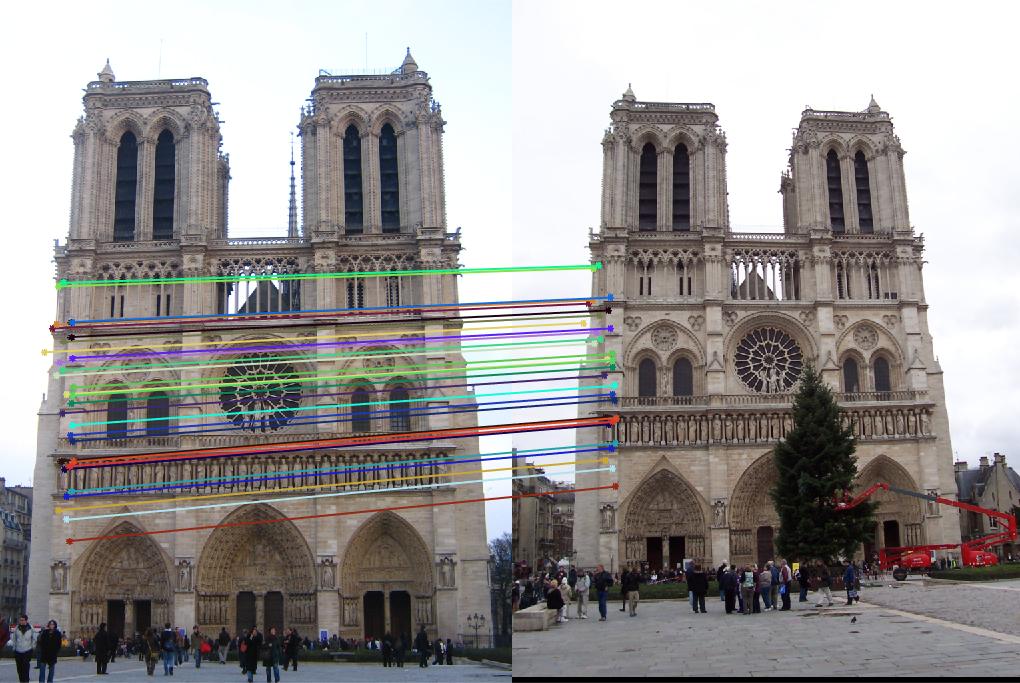
In this part, I set the threshold for counting inliers to be 0.001. The following pictures are results from normalized points and unnormalized points. The normalized one has much more inliers than unnormalized one. Since there are too many points, I set the number of points to display to 30. And I tested 3 pairs of images. The pictures in row 1, 3, 5 are unnormalized, 2, 4, 6 are normalized. It is obvious that the unnormalized one doesn't work well for image Episcopal Gaudi and Notro Dame, the normalized one significantly improved the accuracy and number of inliers. But for Mount Rushmore, the unnormalized one also work good.


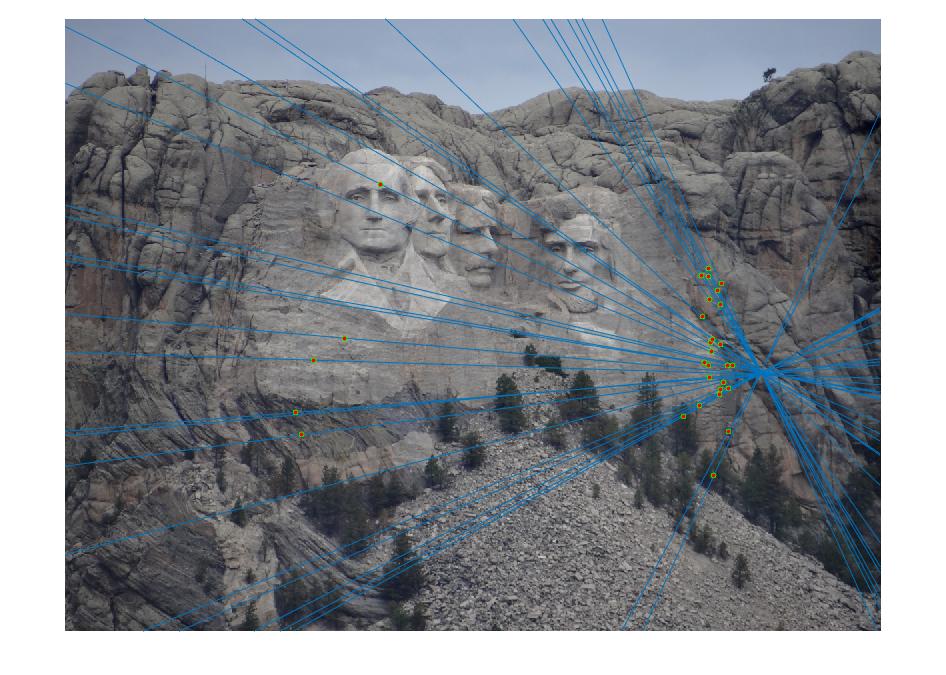
|


|



|



|


|


|
The numbers of inliers from unnormalized and normalized points are shown below. It is obvious that normalized one has more inliers, but it has lower runtime than unnormalized one.
| unnormalized | normalized | |
| Mount Rushmore | 56 | 375 |
| Episcopal Gaudi | 289 | |
| Notre Dame | 118 | 375 |