What is XWRAP Composer?
XWRAP Composer is a semi-automated wrapper generation system that generates wrappers capable of extracting information from multiple heterogeneous Web documents. By encoding wrapper developers' knowledge in Interface Specification, Outerface Specification and Extraction Script, XWRAP Composer integrates single-page wrappers into a composite wrapper capable of extracting information across multiple interconnected pages. The enhancement module allows the generated composite wrapper to be used as a WSDL Web service or a Ptolemy actor for future integration.
Why do we need XWRAP Composer?
Automatically extracting data from Web sites has been an important task for the entire life of the Web. Although there have been many advances in automated information exchange, there remains many example applications that can benefit from automated techniques to extract data from a variety complex Web sites.
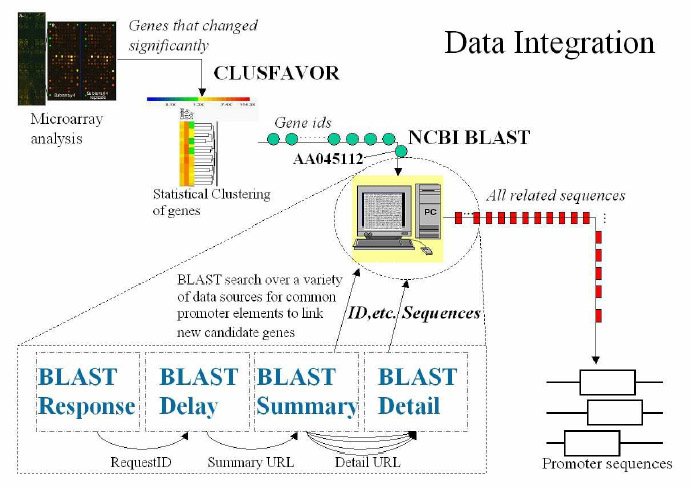
Wrappers generated by wrapper generators, such as XWRAP Original or XWRAP Elite, usually perform well when extracting information from individual documents but are poorly equipped to extract information from multiple linked Web documents. Consider the following typical data integration work flow. A biologist first users a program called Clusfavor to cluster genes that changed significantly in a micro-array analysis experiment. After extracting all gene ids from the Clusfavor result, he feeds them into NCBI Blast Web site, which searches all related sequences over a variety of data sources. The returned sequences will be further examined to find promoter sequences. Each of the phases described above can also be decomposed into smaller tasks. For example, the search in NCBI Blast actually involves four steps. Due to the long waiting time for serving a BLAST search on the NCBI web site, the NCBI Web server will first return a response page with a request ID. When the biologist asks for the results by the request ID, the Web site will presents a delay page if the results are not yet ready to display. Once the search results are delivered, they are displayed in a summary page that contains a summary of all genes matching the search query condition and links to detail pages of each found gene. If the summary page does not include detailed information that the biologist is interested in, he has to visit each detail page.

As usual, a critical challenge for providing system-level support for scientists to achieve such data integration tasks is the problem of locating, accessing, and fusing information from a rapidly growing, heterogeneous, and distributed collection of data sources available on the Web. Automating such complex search and data collection workflow presents three major challenges.
Different websites use different query-answer control logics to present the answer pages to search queries.
Cross-page data extraction has more complex extraction logic than the single page extraction system. Different applications require different sets of data to be extracted by the cross-page data extraction engine. Typically, only portions of one page can the links that lead the extraction to the next page need to be extracted.
Data items extracted from multiple pages require associating with semantically meaningful naming convention, which often provided by the domain scientists who issued such cross-page extraction job.
In answering those challenges, we designed XWRAP Composer, a semi-automated wrapper generation system that generates wrappers capable of extracting information from multiple heterogeneous Web documents.
How does XWRAP Composer toolkit work?
The main idea behind XWRAP Composer toolkit is to separate what to extract from how to extract, and distinguish information extraction logic from query answer control logic. The control logic describes how many different ways a query could be answered from a given web site. The data extraction logic describes the cross page extraction steps, including what information is important to extract at each page and how such information is used as a complex filter in the next search/extraction step. We use interface and outerface description to describe what need to be extracted and user Composer Extraction Script language to describe the control logic and extraction logic and to implement the output alignment and tagging of data items extracted based on the outerface specification.
The following figure presents an architecture sketch of the XWRAP Composer toolkit. XWRAP Composer compiler takes three inputs, interface specification, outerface specification and extraction script, and compiles them into a Java wrapper, which can be further extended into either a Web service with wrapping capability or a Ptolemy wrapper actor for future integration. The compilation process of the XWRAP Composer includes generating code based on control logic and extraction logic as well as generating the correct output alignment and semantically meaningful tagging based on the outerface specification.
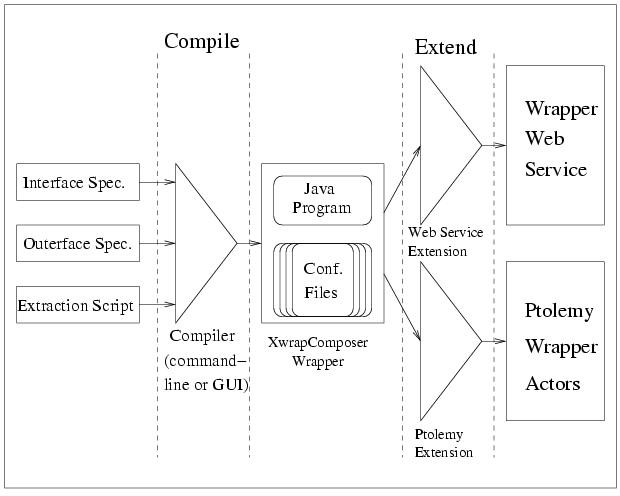
Structure of XWRAP Composer Toolkit
Interface and Outerface Specification.
The design of the XWRAP Composer Interface and Outerface Specification serves two important objectives. First and for most, it will ease the use of XWRAP wrappers. Second, it will also facilitate the XWRAP Composer wrapper code generator system to generate Java code. Therefore, some components of the specification may not be directly useful for the users of these wrappers.
Concretely, the interface specification describes the wrapper name and which web site needs to be wrapped by giving the source URL and other related information. The outerface specification describes what data items are of interest and the semantically meaningful names to be used to tag those data items. The following figure shows a concrete example for the XWRAP Composer interface and outerface schema for the NCBI BLAST summary wrapper.

An example of interface and outerface specification - NCBi Summary
The XWRAP Composer script usually contains three types of root commands, document retrieval, data extraction and post processing. The document retrieval commands construct a file request or an HTTP request and fetch the document. The data extraction commands extract information from the fetched document. The post processing commands allow adding semantic filters to make the output conform to the outerface specification.
The general usage of commands is as follows:

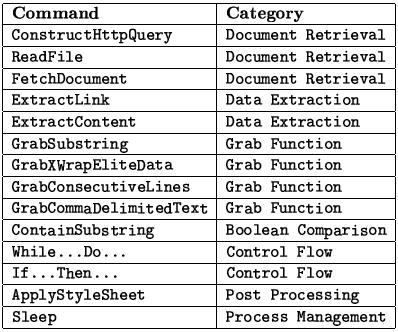
The following table shows a list of commands that are currently supported in the first release of the XWRAP Composer toolkit. Please refer Dr. Wei Han's Ph.D. dissertation for detailed explanation.
The following is an extraction script example for NCBi Summary. Given a sequence from the input, we first construct a NCBi Blast search URL. Script Set variable{[text()]} indicates the sequence is in the input with the XPath, "text()". The first FetchDocument retrieves the NCBi Blast response page that contains a request ID. We extract the ID and construct the URL of search results. The control-flow command while...do... periodically invokes the second FetchDocument to retrieve the result page until the results are delivered. Finally we use GrabXWRAPEliteData to extract useful data from the result page.

Extraction Script Example for NCBi Summary
Code Generation and Extensions
XWRAP Composer compiles interface, outerface and extraction script into an XWRAP Composer wrapper, which contains an executable Java program and a set of configuration files. The configuration files include the input and output schema obtained from interface and outerface, resource files used in the data extraction phase such as XSLT files. Furthermore, because Java programs are usually not convenient for application integration, we have provided two extensions to XWRAP Composer wrappers.
Web services have become a growing trend to access remote applications in data and application integration. In order to make the XWRAP Composer to generate WSDL-enabled wrapper services, one of the extensions we have made to the toolkit is to enable Composer generated wrappers to act as Web services. We carry out this task in two stages. First, we encapsulate an XWRAP Composer wrapper into a general Web service servlet. The servlet automatically extracts the input from a SOAP request, feeds it into the wrapper, and inserts the wrapping results in a SOAP envelope before sending back to the user. Second, WSDL generator can automatically generate Web service description by binding the wrapper's interface and outerface with the servlet configuration. The following figure shows the extensions added to the XWRAP Composer to produce wrappers as WSDL web services.
Ptolemy is a modeling tool to assemble concurrent components, providing a friendly GUI environment to connect task components and govern the interaction between them. The task components have to conform Ptolemy's actor interface in order to be integrated in Ptolemy. We have implemented such an actor interface for generated XWRAP Composer wrappers to be used in Ptolemy. The following two figures demonstrate two ptolemy diagrams that are composed by XWRAP Composer wrapper actors from the left frame.
After clicking on the play button on the top, StartWrapping triggers ReadInputFile to read a gene id from a specified input file. The gene id will then be sent to NCBiSummary Wrapper Actor and the wrapper actor performs the wrapping function upon the gene id input and returns ids of related genes as results. XMLDisplay will pop up a window to present the results.