- February 2021: Aditya is co-organizing the National PREVENT symposium on pandemic prevention and prediction, on behalf of multiple directorates of the NSF. Please join us and register at (or send Aditya an email). We have a terrific set of speakers and participants.
- January 2021: Two papers at AAAI 2021, on disease forecasting using deep learning/machine learning models.
- December 2020: Excited to announce our work (led by PhD student Alex Rodriguez) won 1st place (out of 35 global teams) in the COVID-19 Symptom Data Challenge (organized by Facebook/Catalyst Health/CMU) and 2nd place (out of 777 participants from 43 countries) in the C3AI COVID-19 Grand Challenge! Congratulations to all for this recognition!
- June 2020: Was great particpating in the ML for COVID panel organized by Georgia Tech. Talked about our work in using machine learning for forecasting and surveillance.
- June 2020: Looking forward to leading a new NSF grant (collaborating with UVA and Iowa) on new data-driven methods for Hospital-Acquired-Infections.
- June 2020: New article from Georgia Tech covering our work on CDC COVID forecasting. We are also being covered by Fivethirtyeight.com
- May 2020: A short write-up on our visualization work was released by Berkeley.
- May 2020: Thrilled to be organizing the epiDAMIK 3.0 full-day workshop at ACM SIGKDD 2020, the premier data science conference of the ACM, with colleagues from UVA, Georgetown, Harvard, UIowa and Google. Looking forward to great papers in this extremely timely and crucial area. Papers due June 15. Workshop is on Aug 24.
- May 2020: Congrats to all co-authors, for our AAMAS 2020 paper being selected as one of the 'best papers' of the conference for fast-track publication in JAAMAS.
- April 2020: Congratulations to Dr. Bijaya Adhikari, the newest PhD graduate from our group! Bijaya will join as a tenure-track Asst. Prof. in CS@Iowa in the Fall, after defending a terrific thesis on domain-based frameworks and embeddings for dynamics on networks.
- April 2020: Great to become a member of the modeling infectious diseases MIDAS network.
- March 2020: Thanks to GT CoC for a nice article, discussing some of our past and ongoing work.
- March 2020: Excited to be part of the new NSF CISE Expeditions project on Computational Epidemiology as a Co-PI!
- March 2020: New paper at ACM TIST, on explaining time-series segmentations, joint work with ORNL and VT.
- February 2020: New paper at AAMAS 2020 on designing near-optimal epidemic interventions, to get temporally sensitive policies.


Response
Our Work
Our lab at the Georgia Institute of Technology is participating in multiple activities amidst the ongoing COVID-19 pandemic in the United States, working with partners in academia, industry and public health agencies. We are working on more accurate forecasting models for the CDC, better more adaptive surveillance and testing, easy-to-understand disease visualization overlays and open-source contact tracing efforts. We are also organizing the epiDAMIK workshop at ACM SIGKDD, to bring the data science and public health communities together in this crucial time.
Follow us:News and Articles on our work
1. Forecasting the COVID-19 trajectory
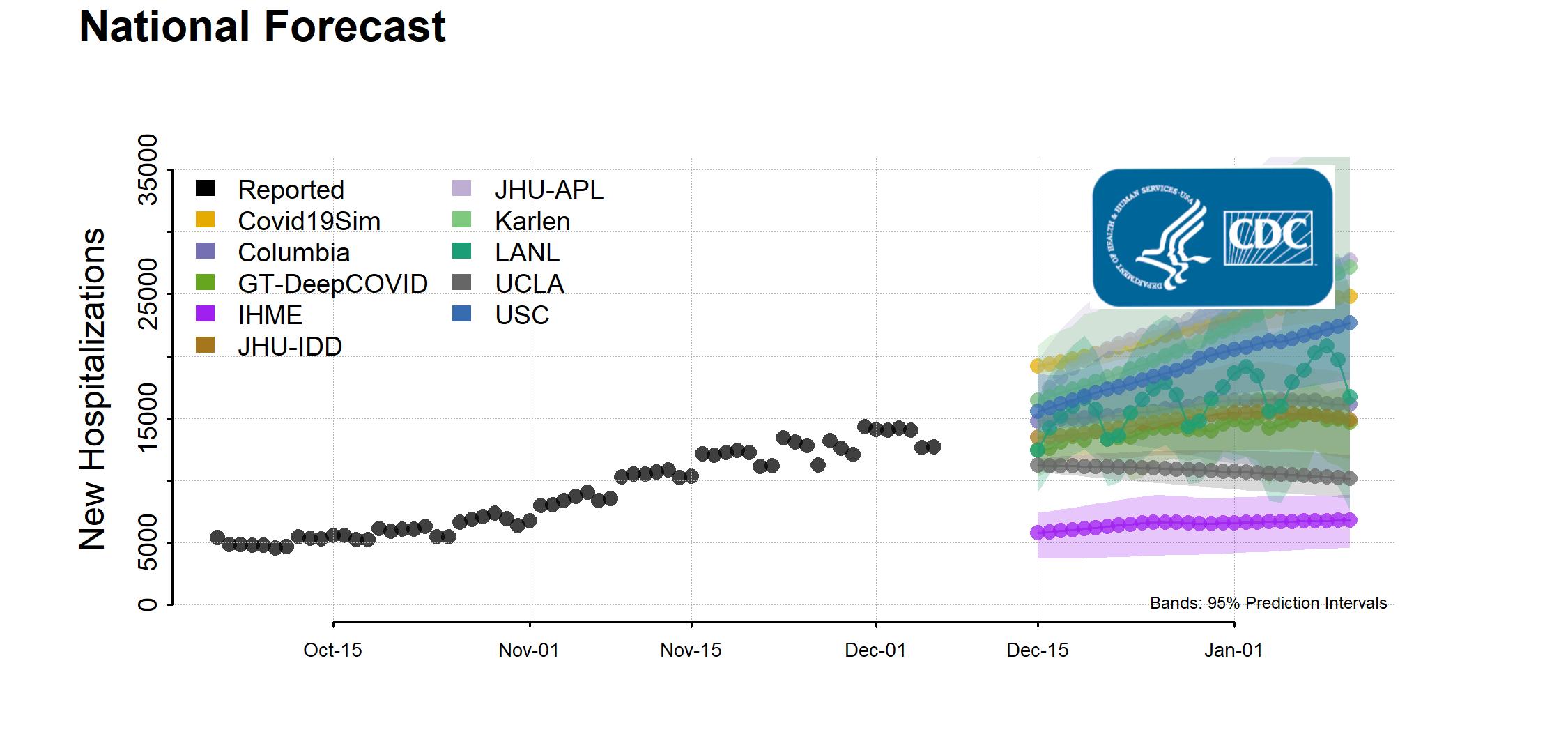
The Centers of Disease Control and Prevention is hosting collaborative forecasting projects related to predicting the coronavirus disease spread, and anticipate the mortality and number of hospitalizations and flu-like-symptoms caused by the disease across the country. This critically timed project comprises a handful of teams of leading data scientists, epidemiologists, statisticians, and high-performance computing researchers from national laboratories, public universities, public health institutions, and some private sector agents. We are leading two teams, one for predicting ILI burden and another for COVID mortality and hospitalizations. Our team is using deep learning models to forecast specific targets at the national, regional, state, and local levels. In addition to CDC ILI and COVID data, we are incorporating many other real-time datasets such as syndromic surveillance data and point-of-care data from major providers. We combine these datasets with domain knowledge using end-to-end deep learning models to predict targets on a weekly basis. The CDC synthesizes our weekly and monthly predictions with other models to help determine policy and other planning decisions to help communities prepare for and fight the disease. We have extensive experience with disease surveillance - we have been leading a team at the CDC FluSight challenge since 2018 (our model EpiDeep had the best performance in the HHS1 region) and have published multiple research papers on incidence prediction at major venues (see related publications below).
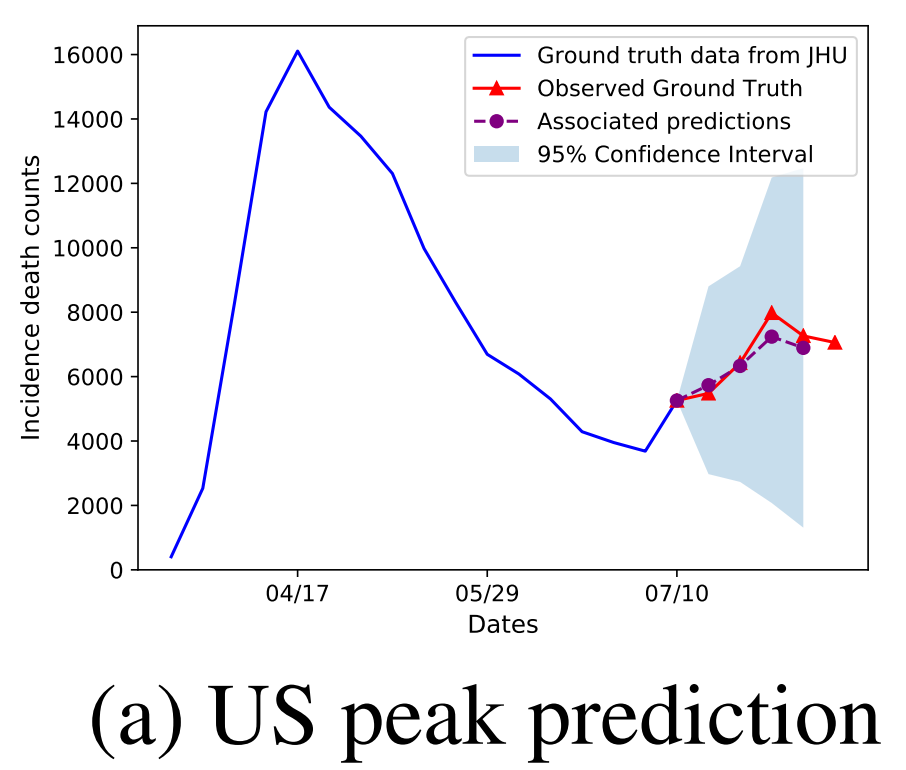
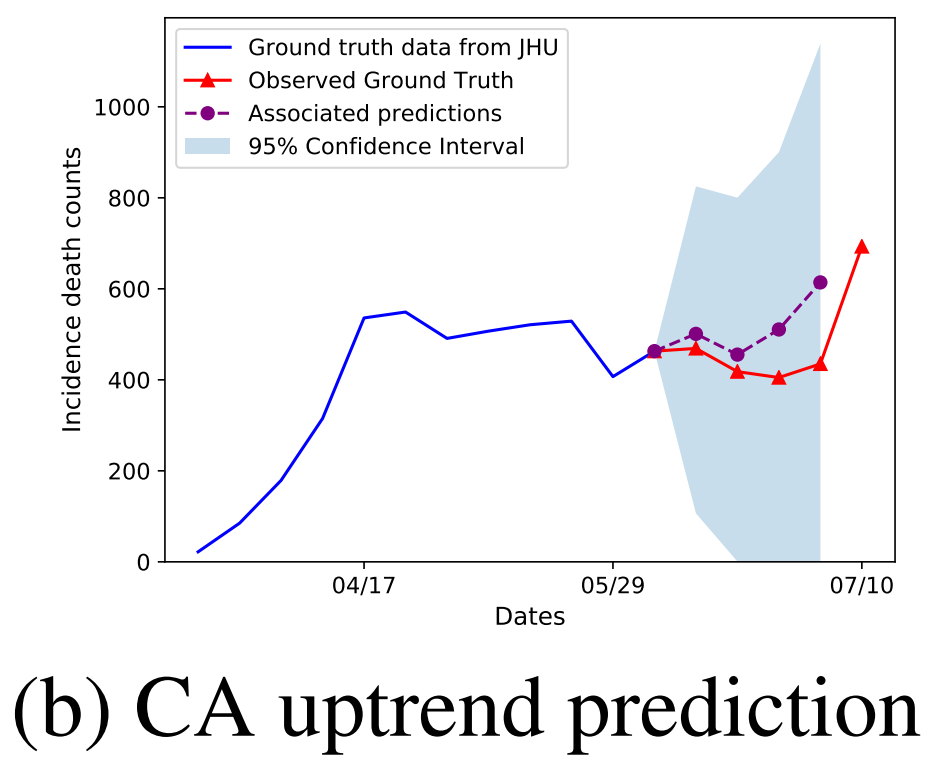
We show two important changes in trend that our model DeepCOVID predicted with several weeks of anticipation: second peak in US national incidence mortality (left) and uptrend in CA incidence mortality (right); both of the pictures above use the JHU dataset as ground truth. Also see our latest results on mortality prediction on the CDC website and FiveThirtyEight (1 out of 11 selected models). Also see our website for DeepCOVID.
- Forecasting COVID-contamined flu (appears in AAAI-21) [PDF]
[CODE][SLIDES].
- DeepCOVID model (appears in IAAI-21) [PDF], appendix [PDF], [SLIDES], and other resources [LINK].
Our team DeepOubreak (led by Georgia Tech, collaborators from UIowa and VT) expanded our published research and participated in several COVID-19 data science challenges. Our work (led by PhD student Alex Rodriguez) was awarded:
- 1st place (out of 35 global teams) in the COVID-19 Symptom Data Challenge (organized by Facebook/Catalyst Health/CMU). See winner announcement on Twitter [LINK], our white paper [PDF] and slides [PDF].
- 2nd place (out of 777 participants from 43 countries) in the C3AI COVID-19 Grand Challenge. See winner announcement on Twitter [LINK] and summary video [LINK].
2. Campus Interventions using WiFI data
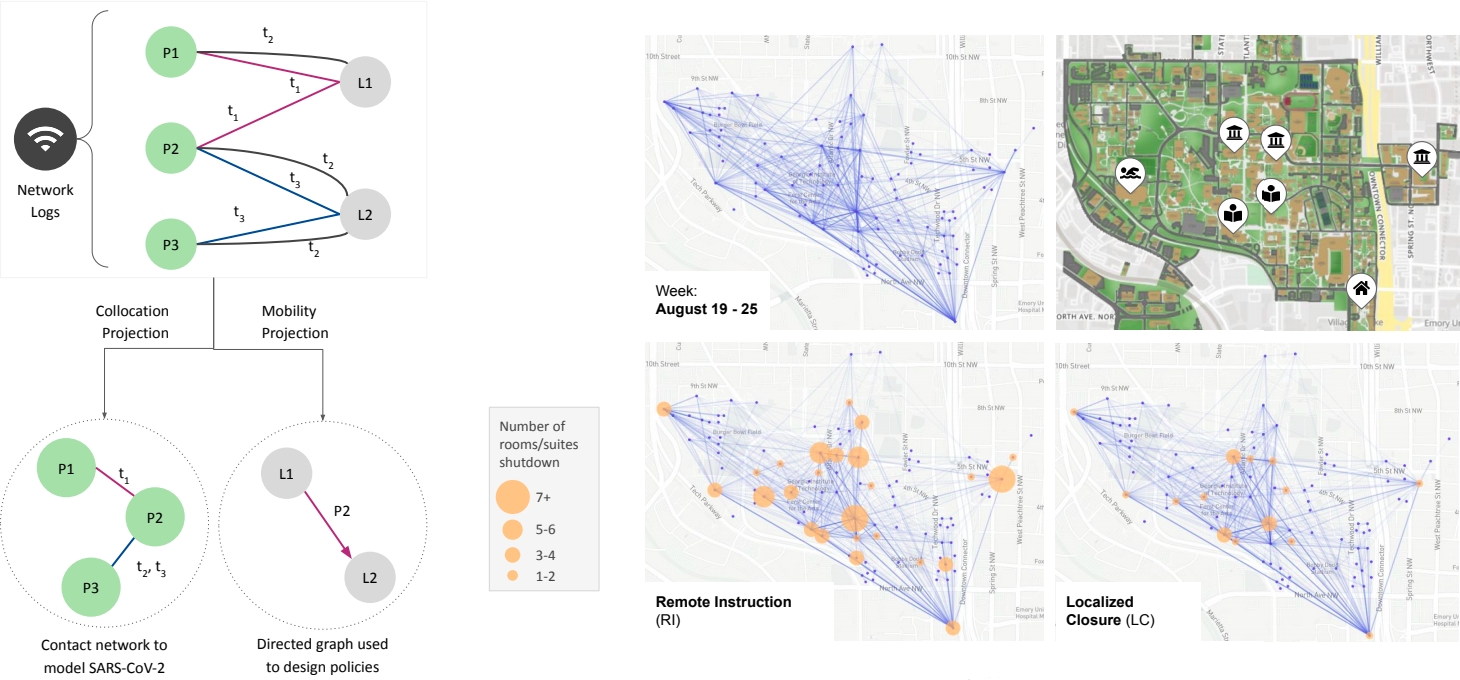
Digital contact tracing is emerging as a possible tool to help with mitigation efforts as societies re-open. We are participating in recent efforts with groups in Georgia Tech and GTRI on both open sourced and close source tools to help make such tracing useful for epidemiolgical modeling.
- Pre-print of WiFi mobility models for campus interventions [PDF].
3. Visualizing the Impact of Non-pharmaceutical Intervention Strategies
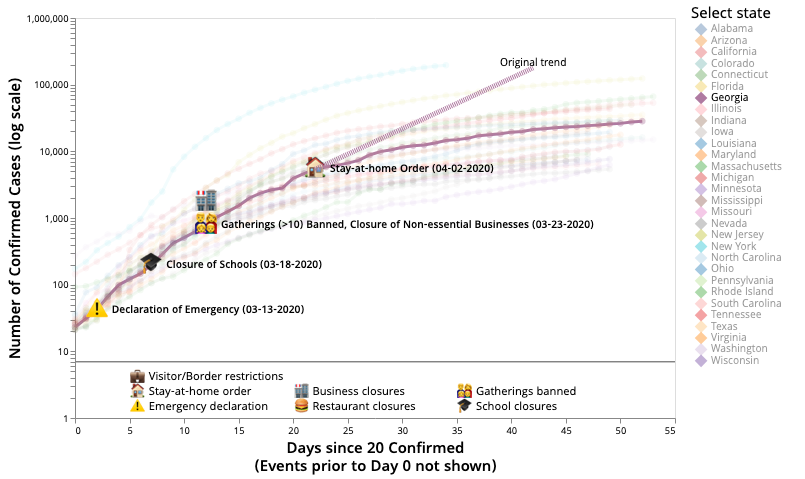
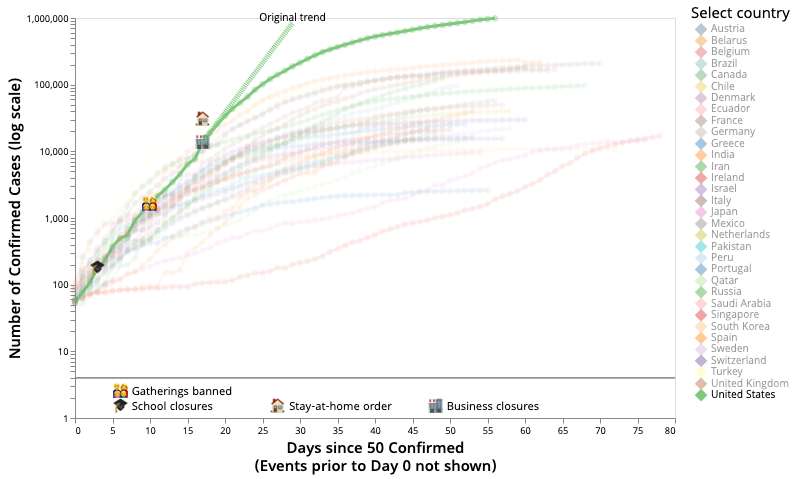
The rapid spread of SARS-CoV-2 has led many countries and regions to enact various interventions, such as social distancing, school closures, and border control, in order to mitigate the growth of infection. Understanding the effects of these interventions is particularly important since each strategy comes with its side effects. We wanted to understand the impact of intervention strategies and their combinations on the disease spread. California Gov. Gavin Newsom mentioned 'Localism is Determinative' for the Covid epidemic. We can clearly see that in our detailed intervention overlays over the disease trajectories of different regions. Our intervention dataset is also public, to help other any computational epidemiology or data science groups. This is a collaboration between data science, computational epidemiology, visualization and public health researchers.
For detailed results and visualizations visit the Covidvis website.
UC Berkeley, BIDS, UIUC, UCSF.
4. Adaptive COVID-19 Surveillance and Testing
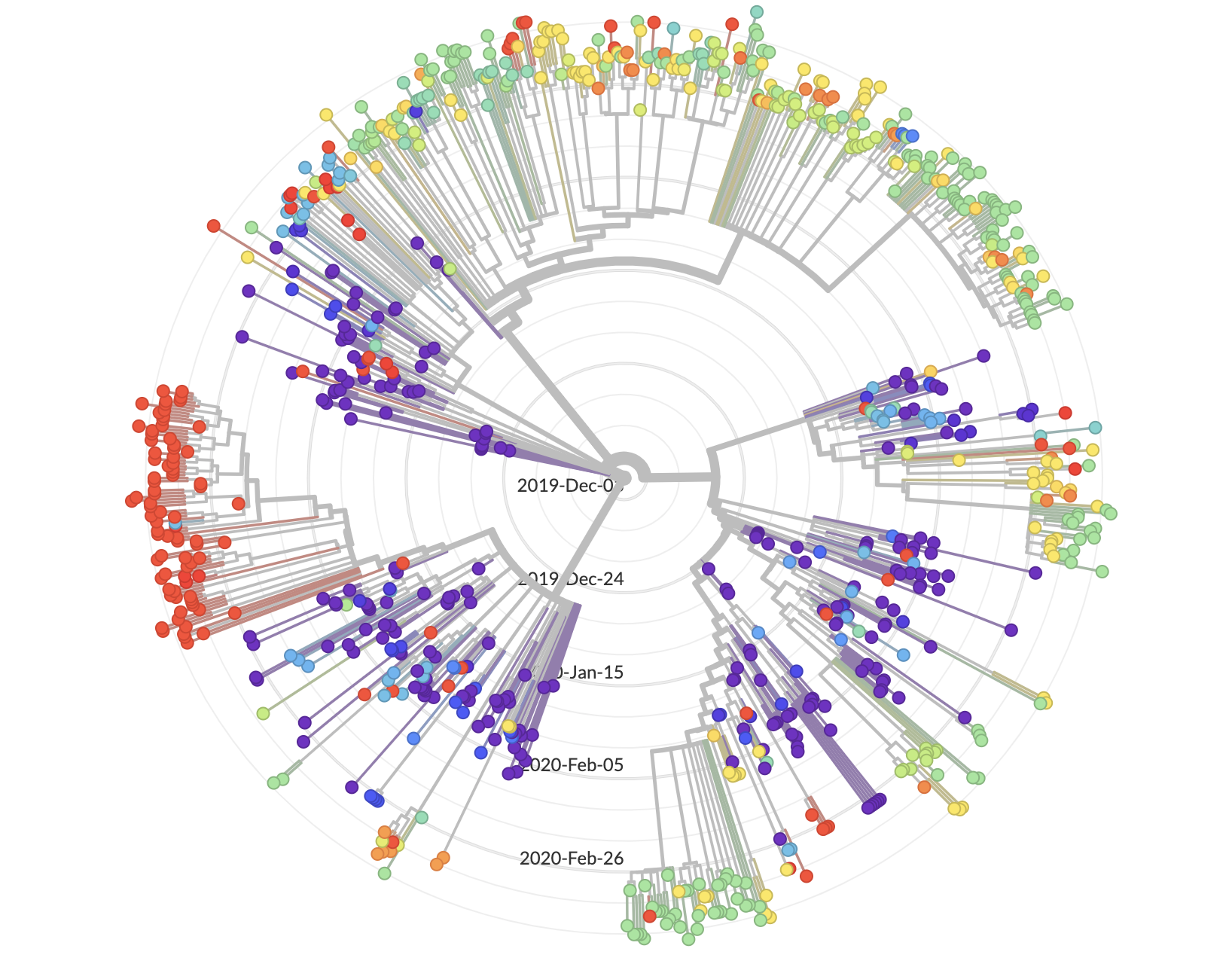
It has been difficult to track and control the COVID-19 pandemic due to various factors such as asymptomatic transmission, high incubation period, human mobility, weather patterns and limited number of tests available. Especially as the number of cases rise, it will become hard to contact-trace everyone, and quarantine appropriately, as experience in other countries shows. Hence, we aim to improve COVID-19 surveillance by more targeted and adaptive testing and intervention in a data-driven fashion. In this newly funded NSF RAPID project, our (a team of Data Science, Network Science, Public Health and Phylogenetic analysis experts) will integrate several datasets including line lists and other auxiliary data like census to answer this question.
For results visit the project website (still under construction).
UVA
5. epiDAMIK 4.0, 'Epi Meets Data' Intl. Workshop at ACM SIGKDD 2021

The devastating impact of the currently unfolding global COVID19 pandemic and those of the Zika, SARS, MERS, and Ebola outbreaks over the past decade has sharply illustrated our enormous vulnerability to emerging infectious diseases. We are living in an era during which human activity is the dominant influence on climate and the environment. With escalating globalization, urbanization, and ecological pressures, the threat of a global pandemic has become more pronounced. There is an urgent need to develop sound theoretical principles and transformative computational approaches that will allow us to address the escalating threat of current and future pandemics. Data mining and Knowledge discovery have an important role to play in this regard. Different aspects of infectious disease modeling, analysis and control have traditionally been studied within the confines of individual disciplines, such as mathematical epidemiology and public health, and data mining and machine learning. Coupled with increasing data generation across multiple domains (like electronic medical records and social media), there is a clear need for analyzing them to inform public health policies and outcomes. Recent advances in disease surveillance and forecasting, and initiatives such as the CDC Flu Challenge, have brought these disciplines closer––public health practitioners seek to use novel datasets and techniques whereas researchers from data mining and machine learning develop novel tools for solving many fundamental problems in the public health policy planning process. We believe the next stage of advances will result from closer collaborations between these two communities---the main objective of epiDAMIK.
Please see here for more details and submission information.
Bijaya Adhikari (UIowa), Rose Yu (UCSD), Amulya Yadav (PennState), Sen Pei (CU), Sarah Kefayati (IBM), Ajitesh Srivastava (USC), Anil Vullikanti (UVA), Arvind Ramanathan (ANL).
6. Nat'l. Symposium on Predicting Emergence of Virulent Entities by Novel Technologies (PREVENT)
In the last year, the ongoing COVID-19 pandemic has severely disrupted the livelihoods of our planet’s human inhabitants, infecting over 85 million individuals, and causing nearly 2 million deaths. What actions should have been taken to minimize the severity of this pandemic (and others before it in the past decades such as Zika, SARS and Ebola)? In retrospect, many actions could have played key roles: environmental monitoring for potential animal-to-human infection spillovers, establishment of pipelines for rapid vaccine development and optimal deployment and distribution, designing data science tools to accurately forecast trajectories, fast and adaptive syndromic surveillance and behavior tracking, designing and timing effective interventions, training susceptible individuals for measures needed to inhibit the spread of infectious agents, and others. What lessons have been learned and what gaps in our knowledge, methodologies, technologies, and policies remain? On behalf of the NSF, we organized a two-day multi-disciplinary National Symposium PREVENT, bringing together experts from various domains to begin to address these and related challenges. This material is based upon work supported by NSF award # 2115126.
Please see here for more details.
Paul Torrens (NYU), Krista Wigginton (Michigan), John Yin (Wisconsin).
Team
- PI: Prof. B. Aditya Prakash, Georgia Tech
- Dr. Bijaya Adhikari, recent PhD from AdityaLab, Asst. Prof. at University of Iowa
- Alexander Rodríguez, CS PhD student at Georgia Tech
- Jiaming Cui, CS PhD student at Georgia Tech
- Anika Tabassum, CS PhD student at Virginia Tech
- Harsha Kamarthi, ML PhD student at Georgia Tech
- Jiajia Xie, MS student at Georgia Tech
- Javen Ho, BS student at Georgia Tech
- Pulak Agarwal, BS student at Georgia Tech
Contact Us
Please contact us with any comments/suggestions. Emails are also welcome if you want to help/collaborate/support our work! We are also always looking out for new interesting problems. Contact information for PI Prakash can be found here.Related Publications
Full list can be found here.- DeepCOVID: An Operational Deep Learning-driven Framework for Explainable Real-time COVID-19 Forecasting [PDF]
Alexander Rodríguez, Anika Tabassum, Jiaming Cui, Jiajia Xie, Javen Ho, Pulak Agarwal, Bijaya Adhikari and B. Aditya Prakash.
in IAAI 2021 (virtual). - Steering a Historical Disease Forecasting Model Under a Pandemic: Case of Flu and COVID-19 [PDF]
Alexander Rodríguez, Nikhil Muralidhar, Bijaya Adhikari, Anika Tabassum, Naren Ramakrishnan and B. Aditya Prakash.
in AAAI 2021 (virtual). - Designing Near-Optimal Temporal Interventions to Contain Epidemics [PDF]
Prathyush Sambaturu, Bijaya Adhikari, B. Aditya Prakash, Srinivasan Venkatramanan, Anil Vullikanti.
in AAMAS 2020, Auckland.
Invited to JAAMAS Special Issue (AAMAS Best papers) - Tracking and Analyzing Dynamics of News-cycles during Global Pandemics: A Historical Perspective [PDF]
Sorour E. Amiri, Anika Tabassum, E. Thomas Ewing and B. Aditya Prakash
in SIGKDD Explorations. Vol 21, Issue 2. 2019. - Fast and Near-Optimal Monitoring for Healthcare Acquired Infection Outbreaks [PDF]
Bijaya Adhikari, Bryan Lewis, Anil Vullikanti, Jose Mauricio Jimenez, and B. Aditya Prakash
in PLoS Computational Biology. 2019. - EpiDeep: Exploiting Embeddings for Epidemic Forecasting [PDF] [CODE]
Bijaya Adhikari, Xinfeng Xu, Naren Ramakrishnan and B. Aditya Prakash
in SIGKDD 2019, Anchorage. - DeepDiffuse: Predicting the 'Who' and 'When' in Cascades [PDF] [CODE]
Mohammad R Islam, Sathappan Muthiah, Bijaya Adhikari, B. Aditya Prakash, and Naren Ramakrishnan
in IEEE ICDM 2018, Singapore. - Data Driven Immunization [PDF] [CODE]
Yao Zhang, Arvind Ramanathan, Anil Vullikanti, Laura Pullum, and B. Aditya Prakash.
in IEEE ICDM 2017, New Orleans.
Invited to KAIS Journal Special Issue (ICDM Best papers) - Reconstructing an Epidemic over Time [PDF] [CODE]
Polina Rozenshtein, Aristides Gionis, B. Aditya Prakash and Jilles Vreeken.
in SIGKDD 2016, San Francisco - Syndromic Surveillance of Flu on Twitter Using Weakly Supervised Temporal Topic Models [PDF] [CODE]
Liangzhe Chen, K. S. M. Tozammel Hossain, Patrick Butler, Naren Ramakrishnan and B. Aditya Prakash
in Data Mining and Knowledge Discovery Journal (DAMI), Springer. 2015.
Support
We thank the following for supporting our lab's work (NSF CAREER, NSF CISE Expeditions, NSF IIS, NSF COVID RAPID, ORNL, GT and GTRI Rapid Seed Funds)


